Publications
View in Pubmed
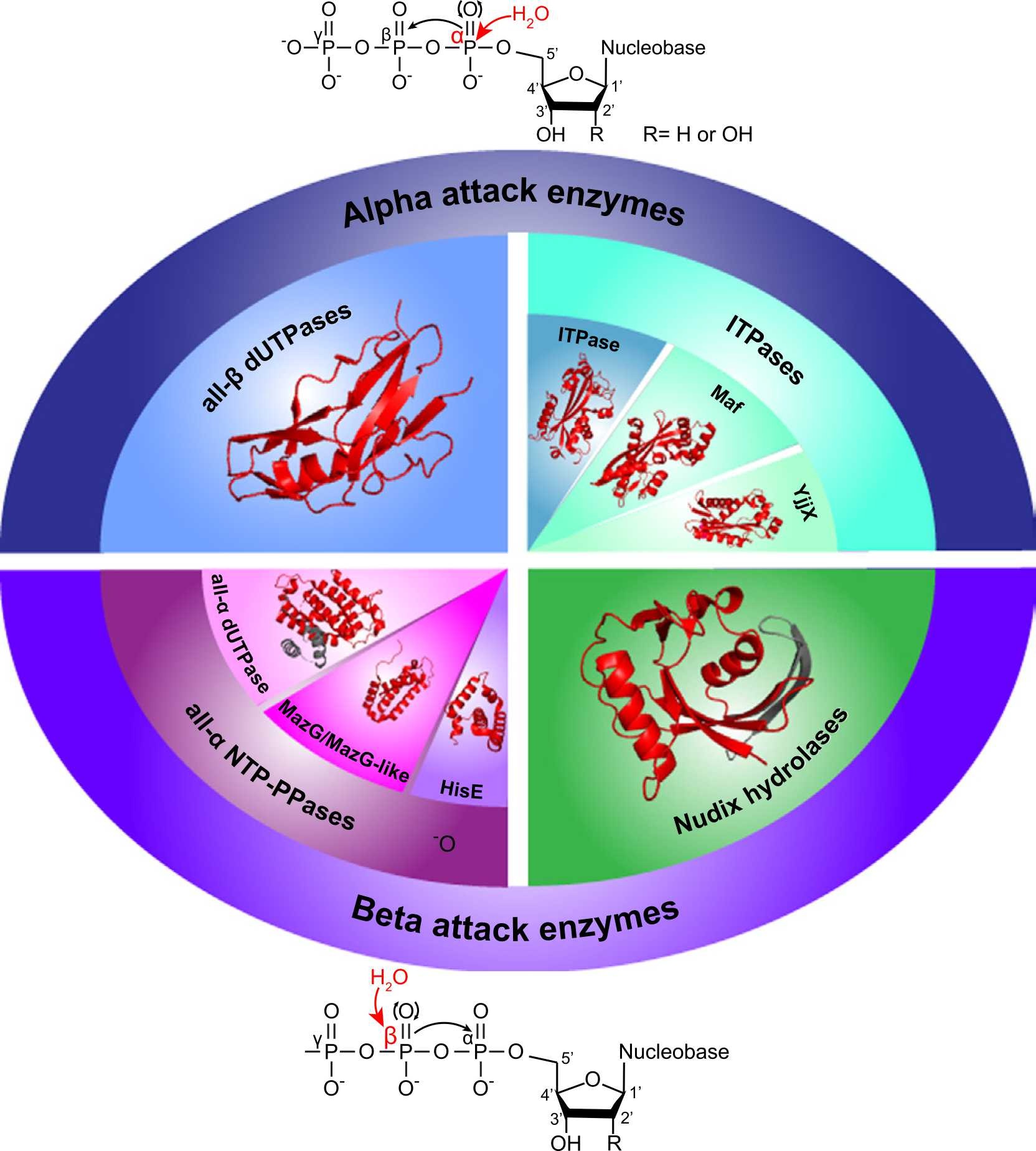
- Sanitation enzymes: Exquisite surveillance of the noncanonical nucleotide pool to safeguard the genetic blueprint
Broderick, K., Moutaoufik, M.T., Aly, K.A., Babu, M.*
Seminars in Cancer Biology 2023 May 19;94:11-20. doi: 10.1016/j.semcancer.2023.05.005. Online ahead of print. (*corresponding)
PMID:37211293
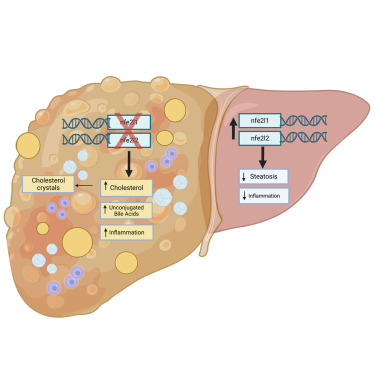
- Complementary gene regulation by NRF1 and NRF2 protects against hepatic cholesterol overload
Akl, M.G., Li, L., Baccetto, R., Phanse, S., Zhang, Q., Trites, M.J., McDonald, S., Aoki, H., Babu, M., Widenmaier, S.B.
Cell Reports 2023 Apr 14;42(4):112399. doi: 10.1016/j.celrep.2023.112399
PMID:337060561
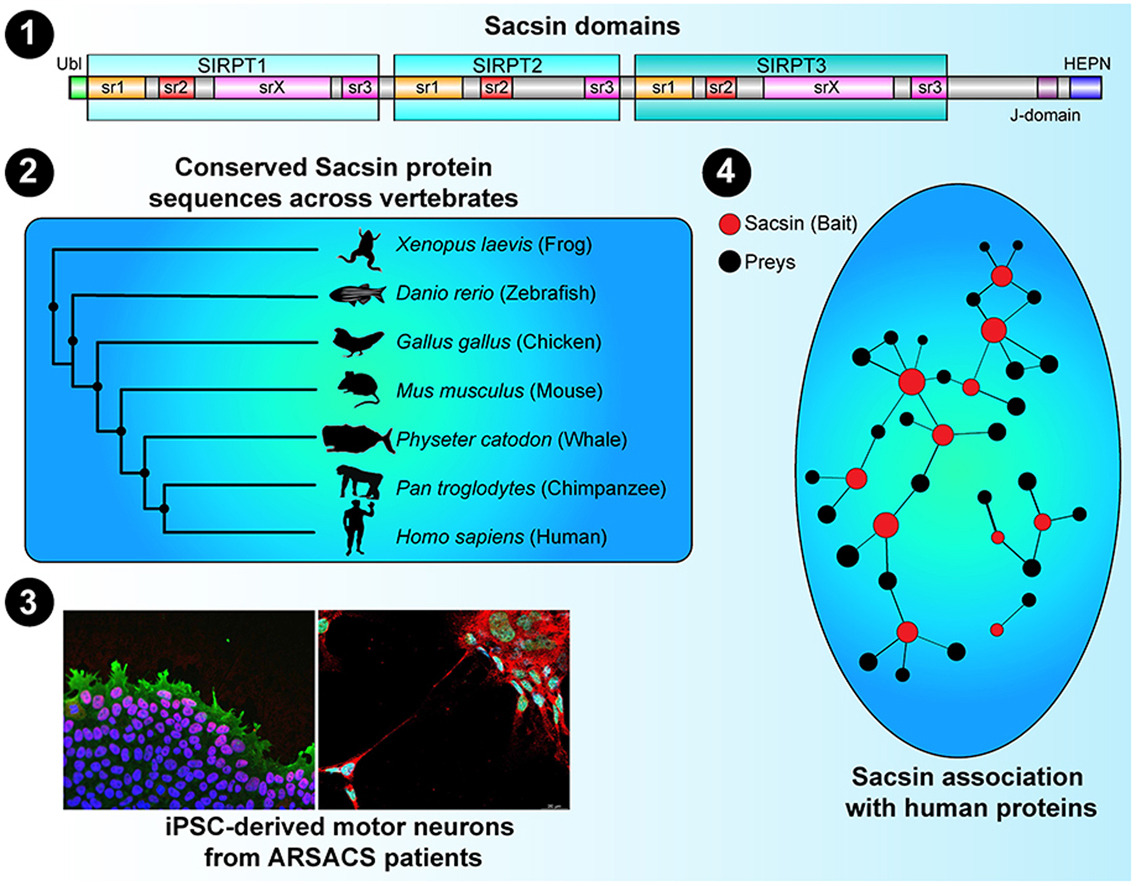
- Insights into SACS pathological attributes in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS)☆
Aly, K.A., Moutaoufik, M.T., Zilocchi, M., Phanse, S., Babu, M.*
Current Opinion Chemical Biology. 2022 Sep 17;71:102211. (*corresponding)
PMID:36126381
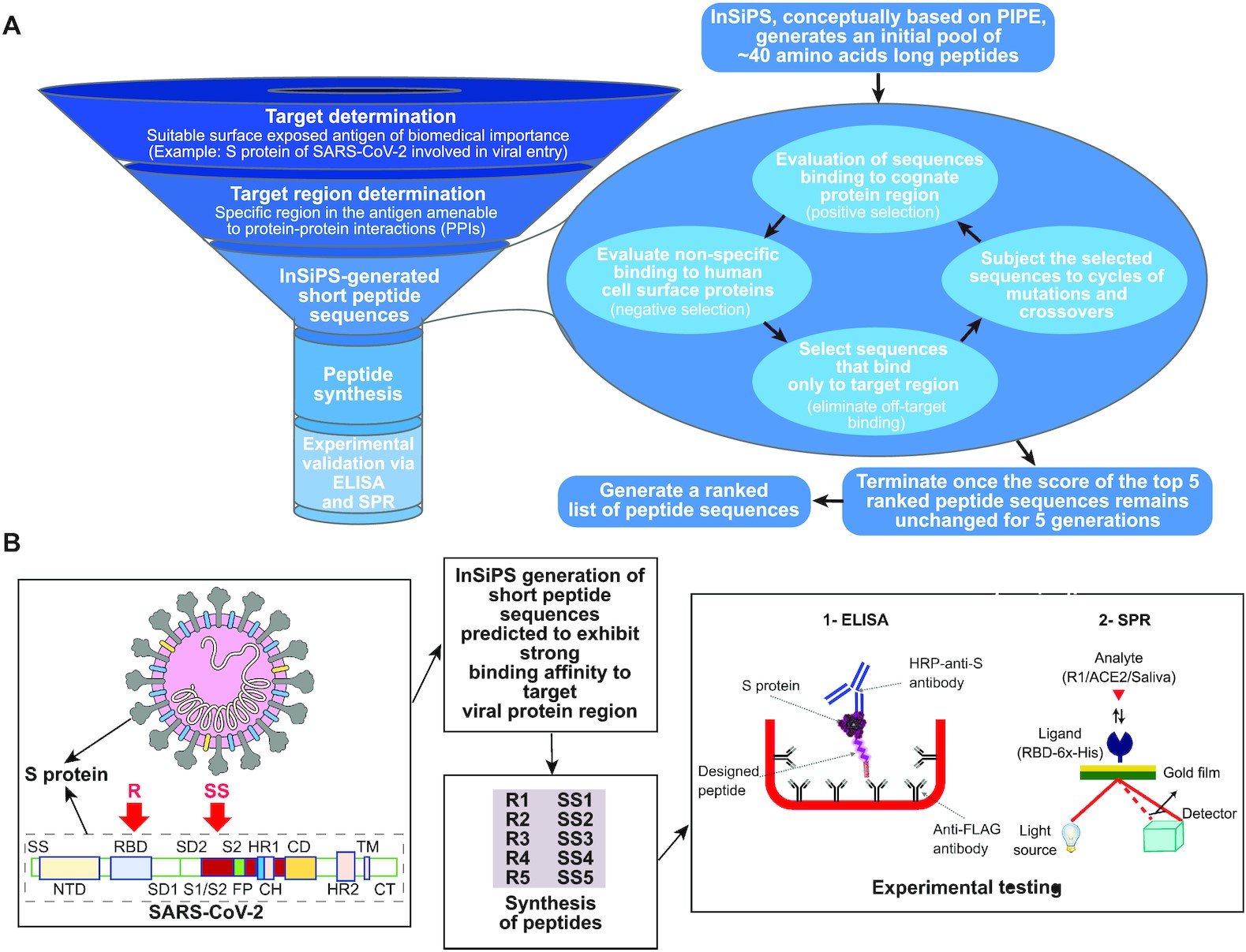
- A computational approach to rapidly design peptides that detect SARS-CoV-2 surface protein S
Hajikarimlou, H., Hooshyar, M., Moutaoufik, M.T., Aly, K.A., Azad, T., Takallou, S., Jagadeesan, S., Phanse, S., Said, K.B., Samanfar, B., Bell, J.C., Dehne, F., Babu, M.*, Golshani, A.
NAR Genomics and Bioinformatics, Volume 4, Issue 3, September 2022, lqac058 (*corresponding)
PMID:36004308
- Auxotrophic and prototrophic conditional genetic networks reveal the rewiring of transcription factors in Escherichia coli.
Gagarinova, A., Hosseinnia, A., Rahmatbakhsh, M., Istace, Z., Phanse, S., Moutaoufik, M.T., Zilocchi, M., Zhang, Q., Aoki, H., Jessulat, M., Kim, S., Aly, K.A., & Babu, M.*
Nature Communications 2022 Jul 14;13(1):4085 (*corresponding)
PMID:35835781
- CFTR interactome mapping using the mammalian membrane two-hybrid high-throughput screening system
Lim, S.H., Snider, J., Birimberg-Schwartz, L., Ip, W., Serralha, J.C., Botelho, H.M., Lopes-Pacheco, M., Pinto, M.C, Moutaoufik, M.T., Zilocchi, M., Laselva, O., Esmaeili, M., Kotlyar, M., Lyakisheva, A., Tang, P., Vázquez, L.L., Akula, I., Aboualizadeh, F., Wong, V., Grozavu, I., Opacak-Bernardi, T., Yao, Z., Mendoza. M., Babu, M., Jurisica, I., Gonska, T., Bear, C.E., Amaral, M.D., Stagljar, I.
Molecular Systems Biology 2022 Feb;18(2):e10629.
PMID:35156780
- Adenosine A1 receptor ligands bind to α-synuclein: implications for α-synuclein misfolding and α-synucleinopathy in Parkinson's disease
Jakova, E., Moutaoufik, M.T., Lee, J.S., Babu, M., Cayabyab.F.S.
Translational Neurodegenertion 2022 Feb 10;11(1):9.
PMID:35139916
- Assembly principles of the human R2TP chaperone complex reveal the presence of R2T and R2P complexes
Seraphim, T.V., Nano, N., Cheung, Y.W.S., Aluksanasuwan, S., Colleti, C., Mao, Yu-Q., Bhandari, V., Young, G., Höll, L., Phanse, S., Gordiyenko, Y., Southworth, D.R., Robinson, C.V., Thongboonkerd, V., Gava, L.M., Borges, J.C., Babu, M., Barbosa, L.R.S., Ramos, C.H.I., Kukura, P., Houry, W.A.
Structure . 2022 Jan 6;30(1):156-171.e12.
PMID:34492227
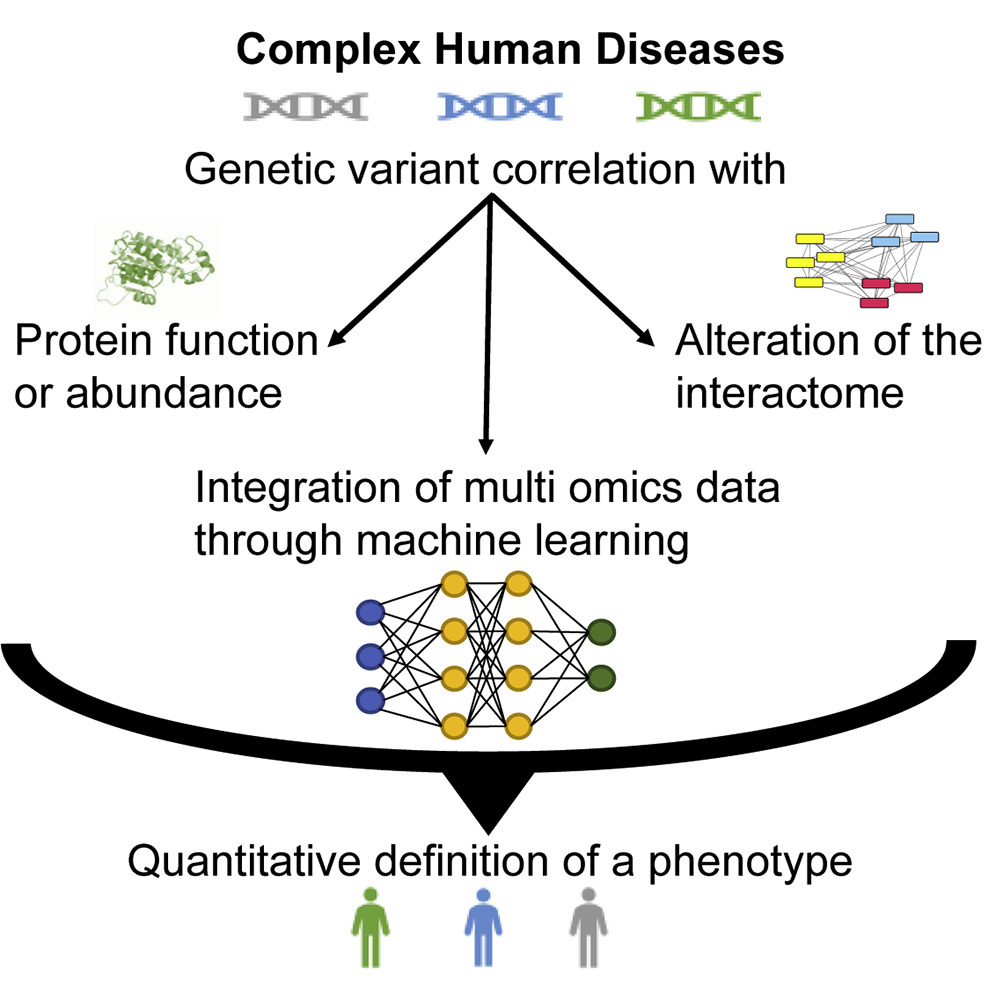
- A Panoramic View of Proteomics and Multiomics in Precision Health
Zilocchi, M., Wang, C., Babu, M.*, Li, J.*
iScience, 2021 Jul 30;24(8) (*co-corresponding)
PMID:34430814
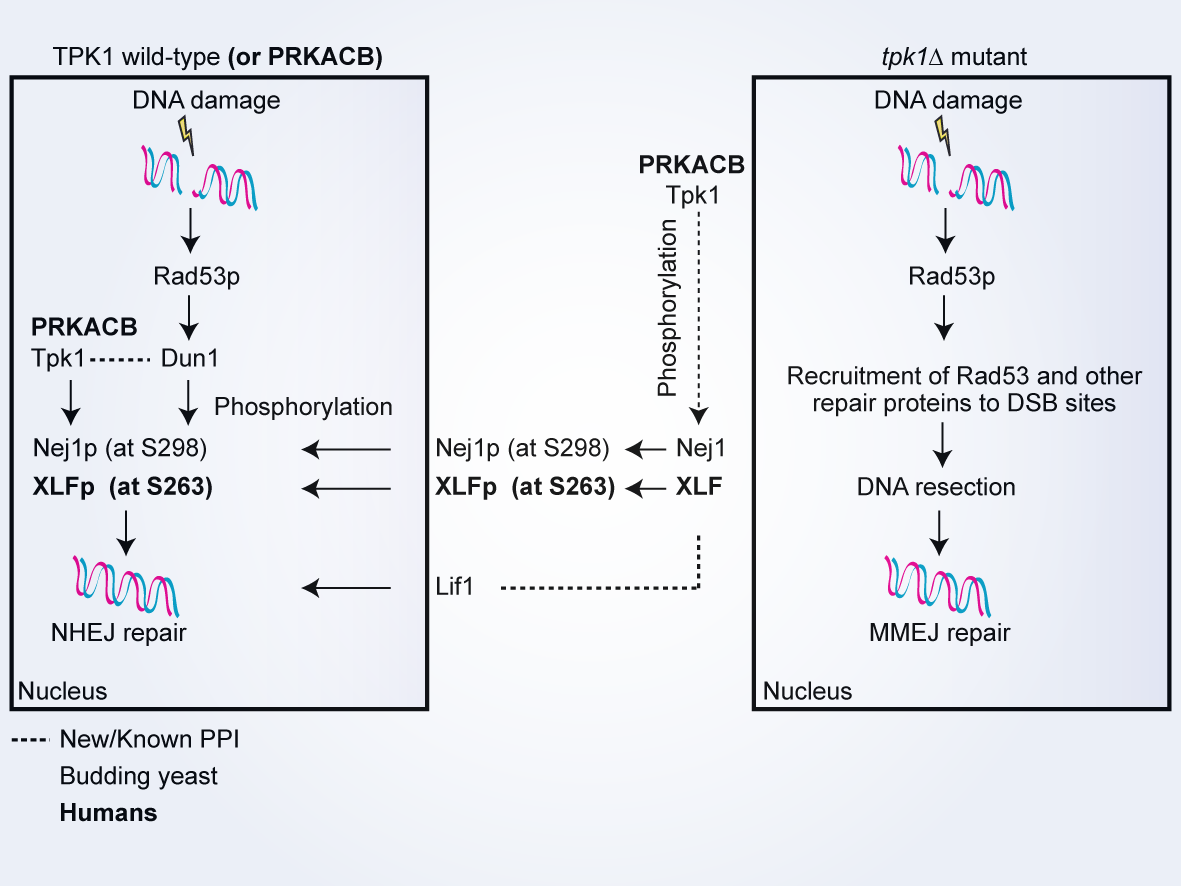
- The conserved Tpk1 regulates non-homologous end joining double-strand break repair by phosphorylation of Nej1, a homolog of the human XLF
Jessulat, M., Amin, S., Hooshyar, M., Malty, R., Moutaoufik, M.T., Zilocchi, M., Istace, Z., Phanse, S., Aoki, H., Omidi, K., Burnside, D., Samanfar, B., Aly, K.A., Golshani, A., Babu, M.*
Nucleic Acids Research, 2021 Aug 20;49(14):8145-8160 (*corresponding)
PMID:34244791
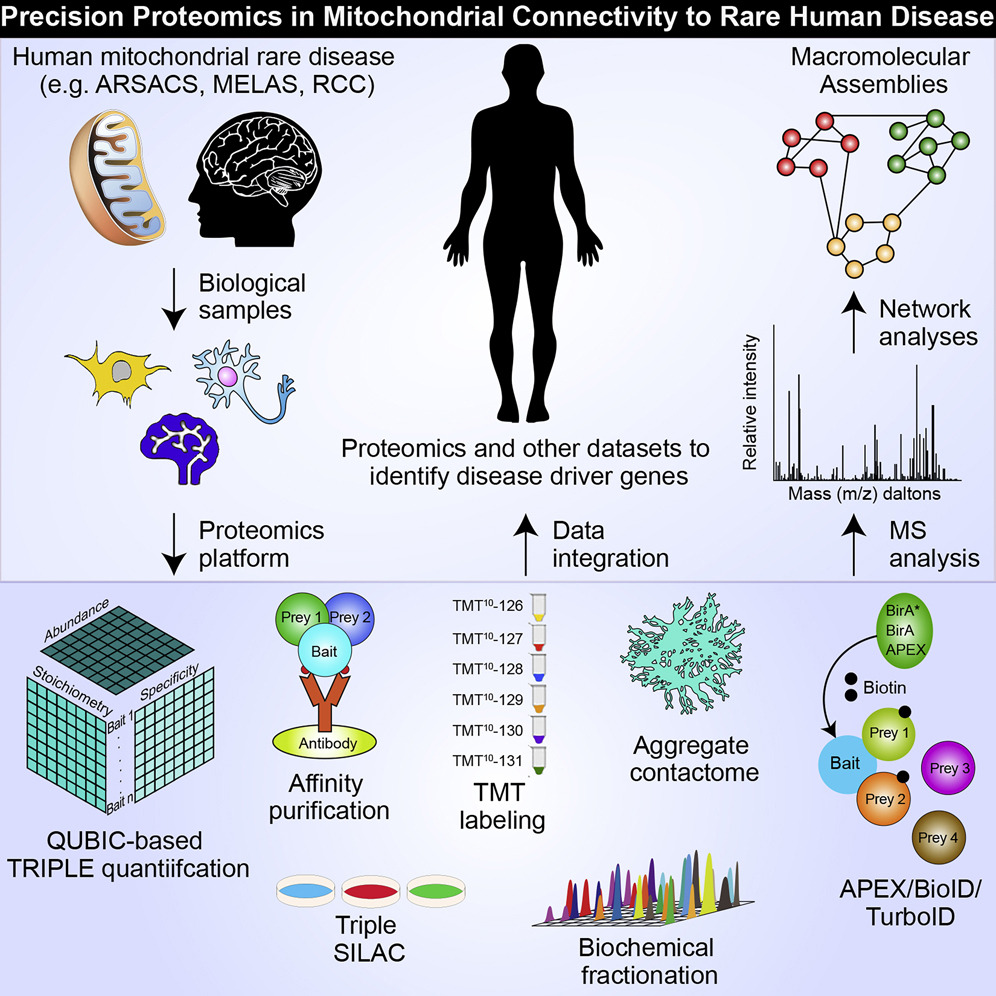
- From fuzziness to precision medicine: on the rapidly evolving proteomics with implications in mitochondrial connectivity to rare human disease
Aly, K.A., Moutaoufik, M.T., Phanse, S., Zhang, Q., Babu M.*
iScience 2021;24(2). (*corresponding)
PMID:33521598
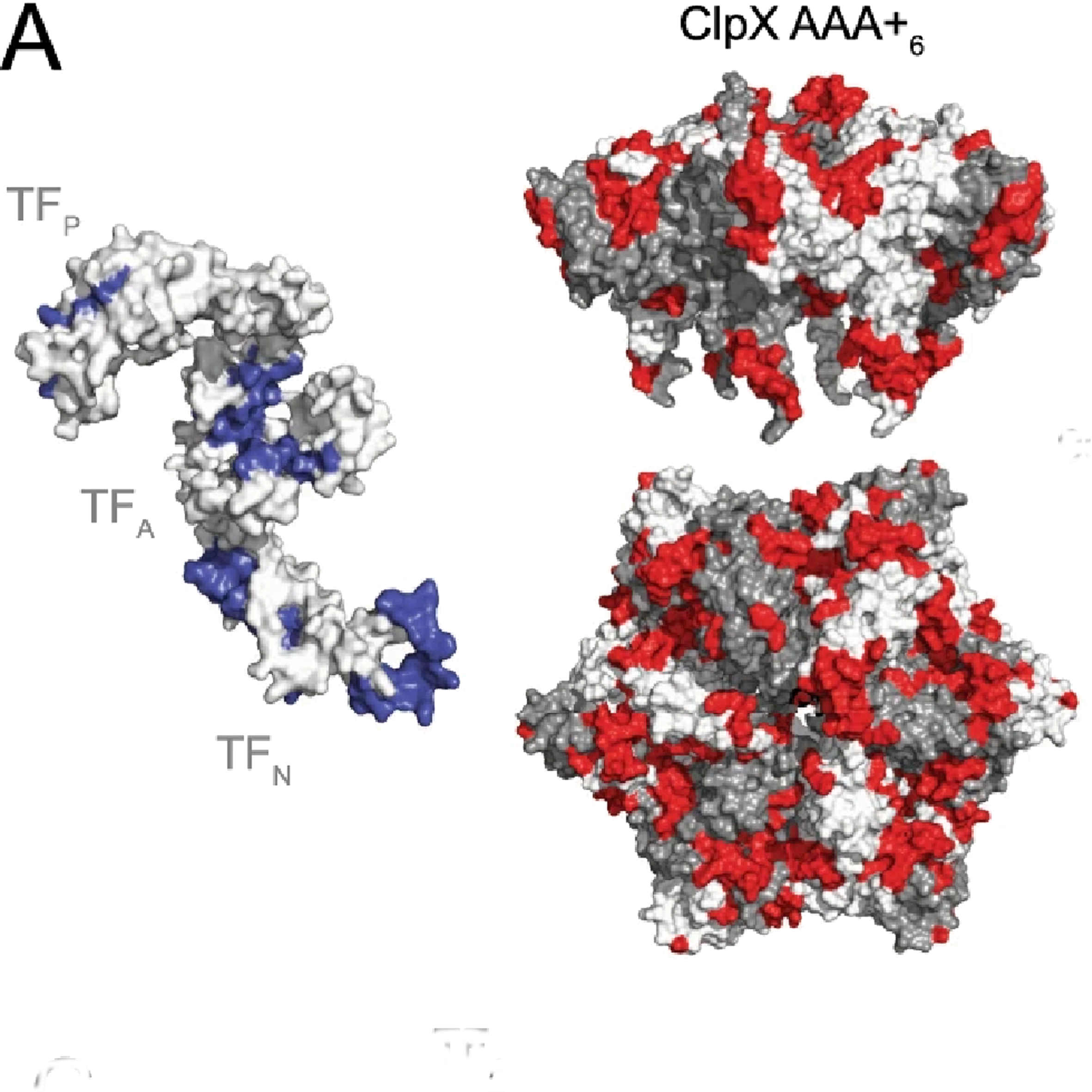
- Functional cooperativity between the trigger factor chaperone and the ClpXP proteolytic complex
Rizzolo, K., Yu, A. H. Y., Ologbenla, A., Kim, S., Zhu, H., Ishimori, K., Thibault, G., Leung, E., Zhang, Y.W., Teng, M., Haniszewski, M., Miah, N., Phanse, S., Minic, Z., Lee, S., Caballero, J.D., Babu, M., Tsai, F.T.F., Saio T., & Houry, W.A.
Nature Communications 2021;12(1):281.
PMID:33436616
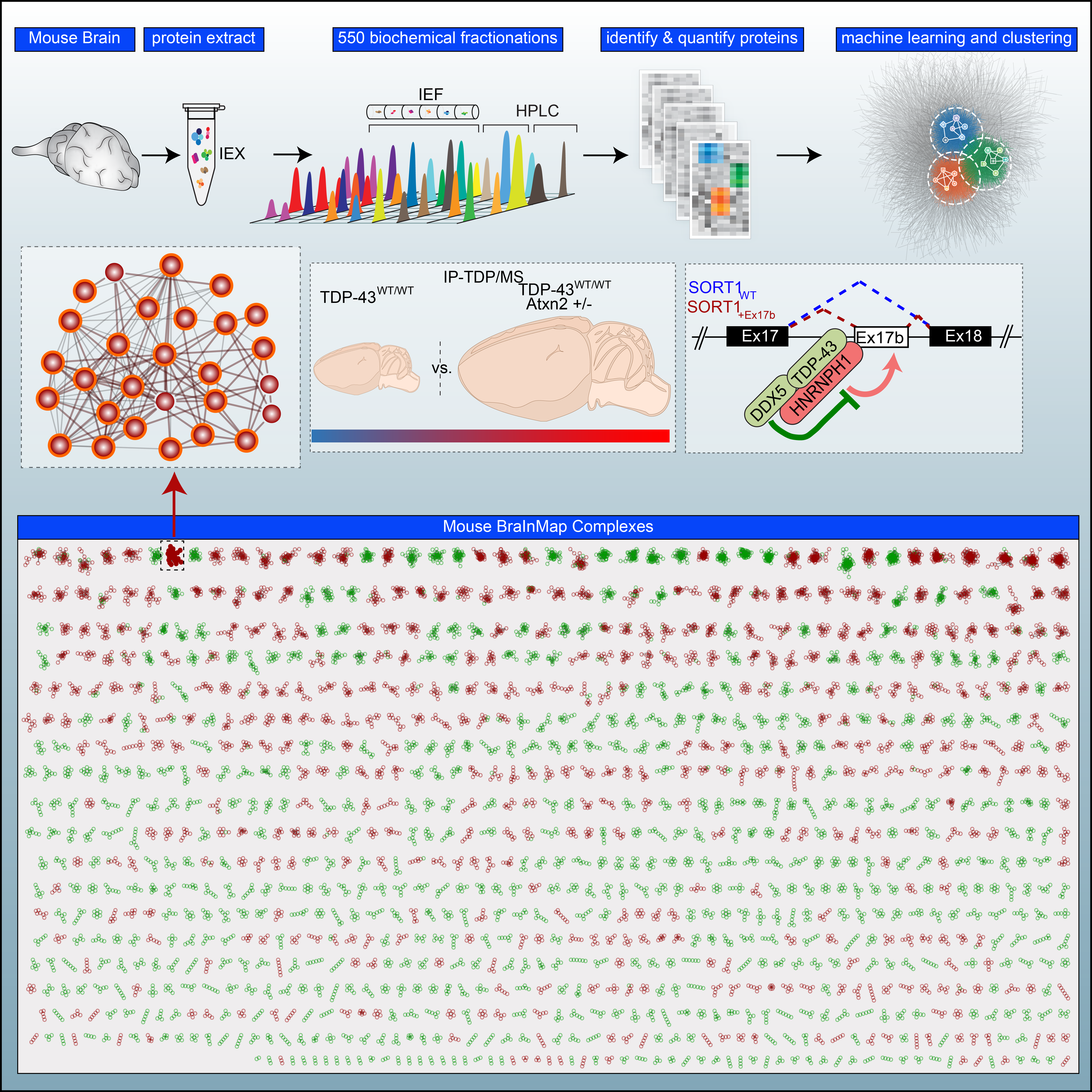
- BraInMap Elucidates the Macromolecular Connectivity Landscape of Mammalian Brain
Pourhaghighi, R., Ash, P.E.A., Phanse, S., Goebels, F., Hu, L.Z.M., Chen, S., Zhang, Y., Wierbowski, S.D., Boudeau, S., Moutaoufik, M.T., Malty, R.H., Malolepsza, E., Tsafou, K., Nathan, A., Cromar, G., Guo, H., Abdullatif, A.A., Apicco, D.J., Becker, L.A., Gitler, A.D., Pulst, S.M., Youssef, A., Hekman, R., Havugimana, P.C., White, C.A., Blum, B.C., Ratti, A., Bryant, C.D., Parkinson, J., Lage, K., Babu, M., Yu, H., Bader, G.D., Wolozin, B., Emili, A.
Cell Systems 2020;10(4):333-350.e14.
PMID:32325033
- HERG channel and cancer: A mechanistic review of carcinogenic processes and therapeutic potential
He, S., Moutaoufik, M.T., Islam, S., Persad, A., Wu, A., Aly, K.A., Fonge, H., Babu, M., Cayabyab, F.S.
Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 2020;1873(2):188355.
PMID:32135169
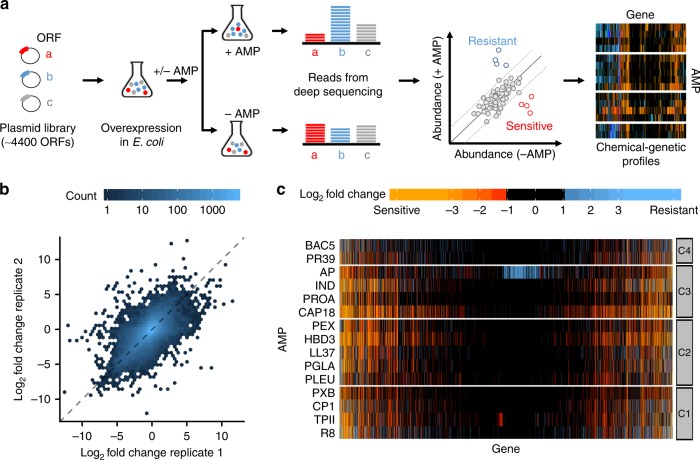
- Chemical-genetic profiling reveals limited cross-resistance between antimicrobial peptides with different modes of action
Kintses, B., Jangir, P.K., Fekete, G., Számel, M., Méhi, O., Spohn, R., Daruka, L., Martins, A., Hosseinnia, A., Gagarinova, A., Kim, S., Phanse, S., Csörgő, B., Györkei, A., Ari, E., Lázár, V., Nagy, I., Babu, M., Pál C., & Papp, B.
Nature Communications 2019;10(1):5731.
PMID:31844052
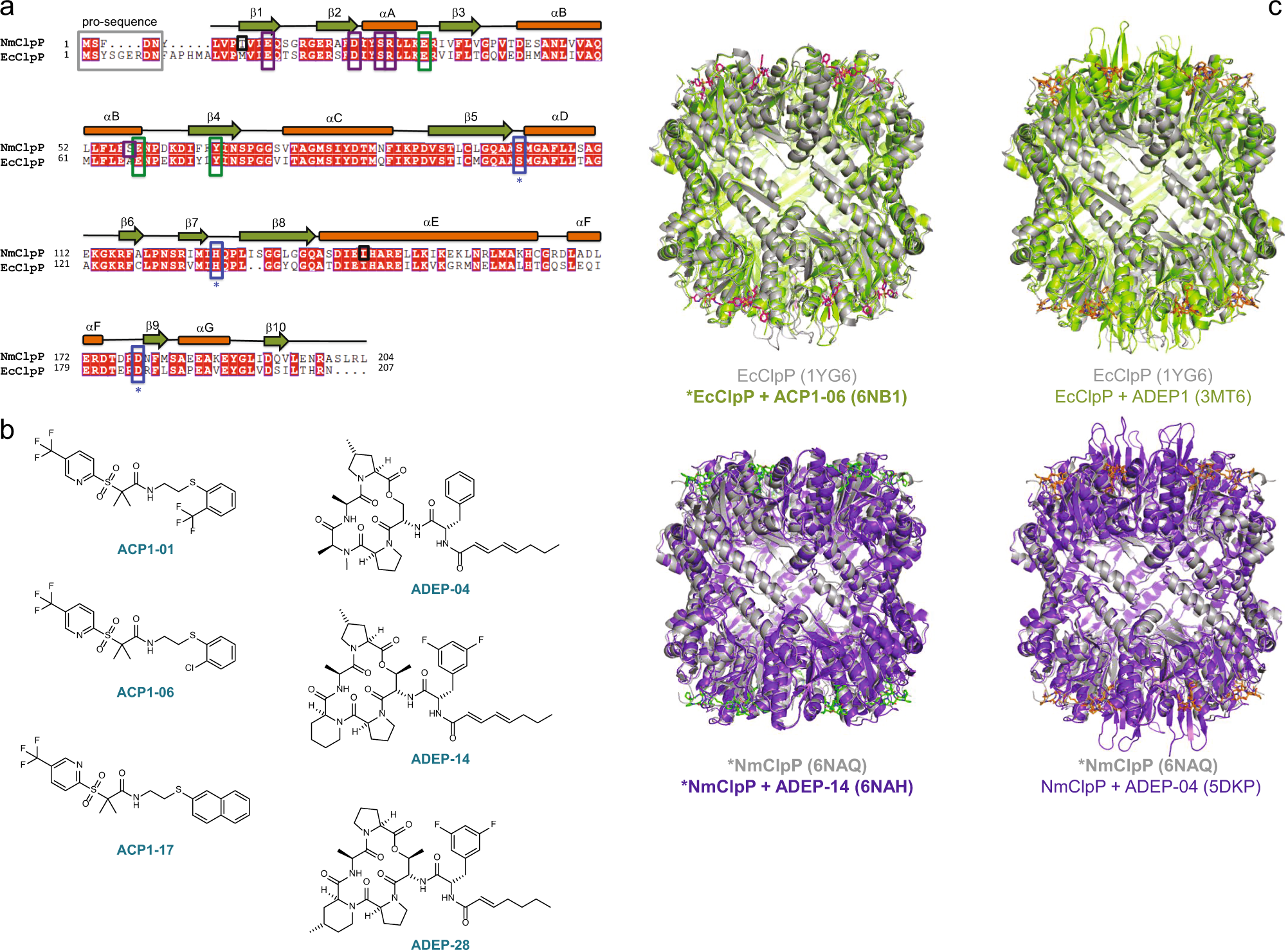
- ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores
Mabanglo, M.F., Leung, E., Vahidi, S., Seraphim, T.V., Eger, B.T., Bryson, S., Bhandari, V., Zhou, J.L., Mao, Y., Rizzolo, K., Barghash, M.M., Goodreid, J.D., Phanse, S., Babu, M., Barbosa, L.R.S., Ramos, C.H.I., Batey, R.A., Kay, L.E., Pai, E.F., Houry, W.A.
Communications Biology - Nature 2019;2:410.
PMID:31754640
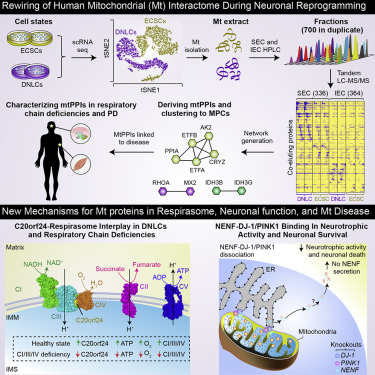
- Rewiring of the Human Mitochondrial Interactome During Neuronal Reprogramming Reveals Regulators of the Respirasome and Neurogenesis
Moutaoufik, M.T., Malty, R., Amin, S., Zhang, Q., Phanse, S., Gagarinova, A., Zilocchi, M., Hoell, L., Minic, Z., Gagarinaova, M., Aoki, H., Stockwell, J., Jessulat, M., Goebels, F., Broderik, K., Scott, N.E., Vlasblom, J., Musso, G., Prasad, B., Lamantea, E., Garavaglia, B., Rajput, A., Murayama, K., Okazaki, Y., Foster, L.J., Bader, G.D., Cayabyab, F.S., Babu, M.*
iScience 2019;19:1114-1132. (*corresponding)
PMID:31536960
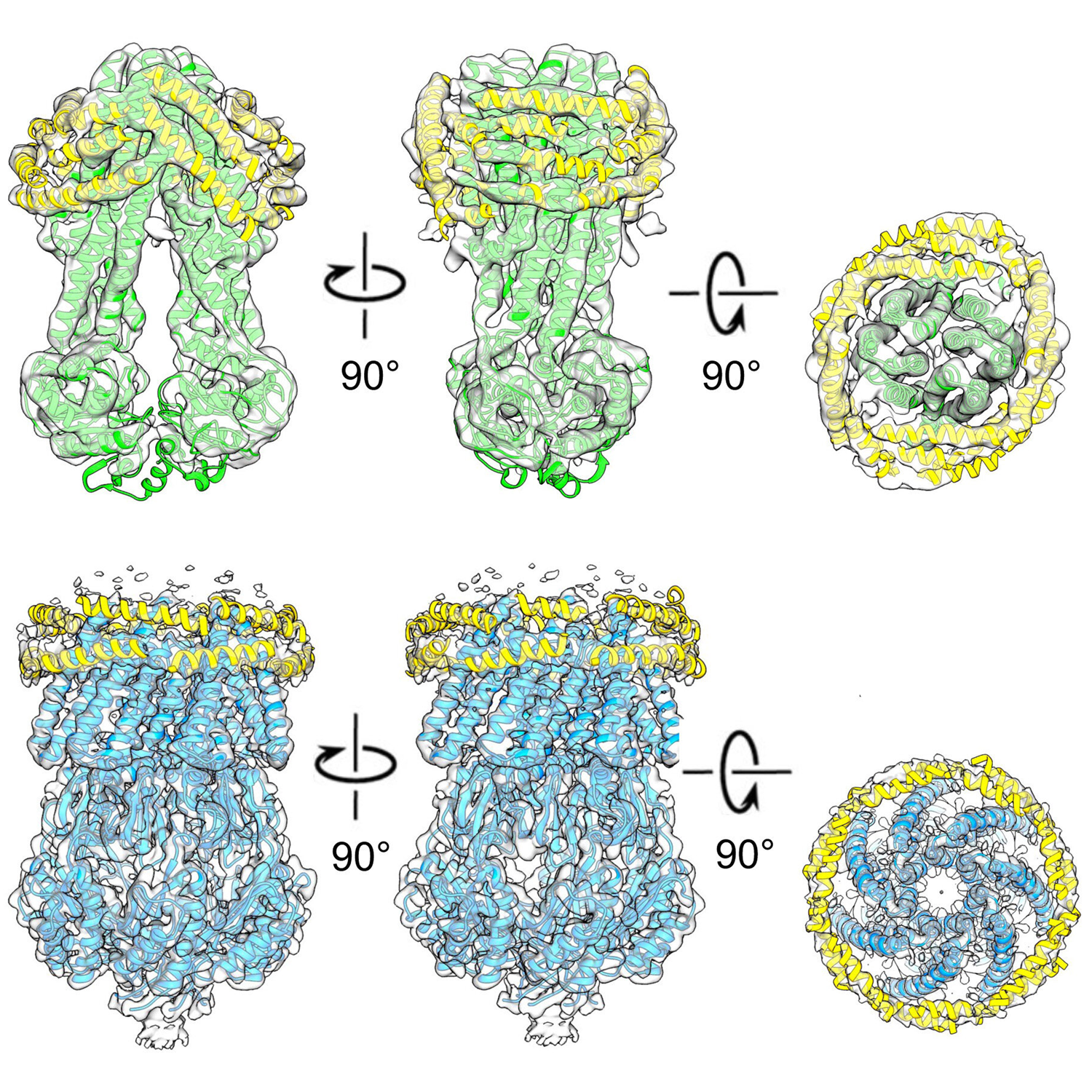
- Profiling the Escherichia coli membrane interactome captured in peptidisc libraries
Carlson, M.L., Stacey, R.G., Young, J.W., Wason, I.S.,Zhao, Z., Rattray, D.G., Scott, N., Kerr, C.H., Babu, M., Foster, L.J., Duong F.V.H.
eLife 2019;8. pii: e46615.
PMID:31364989
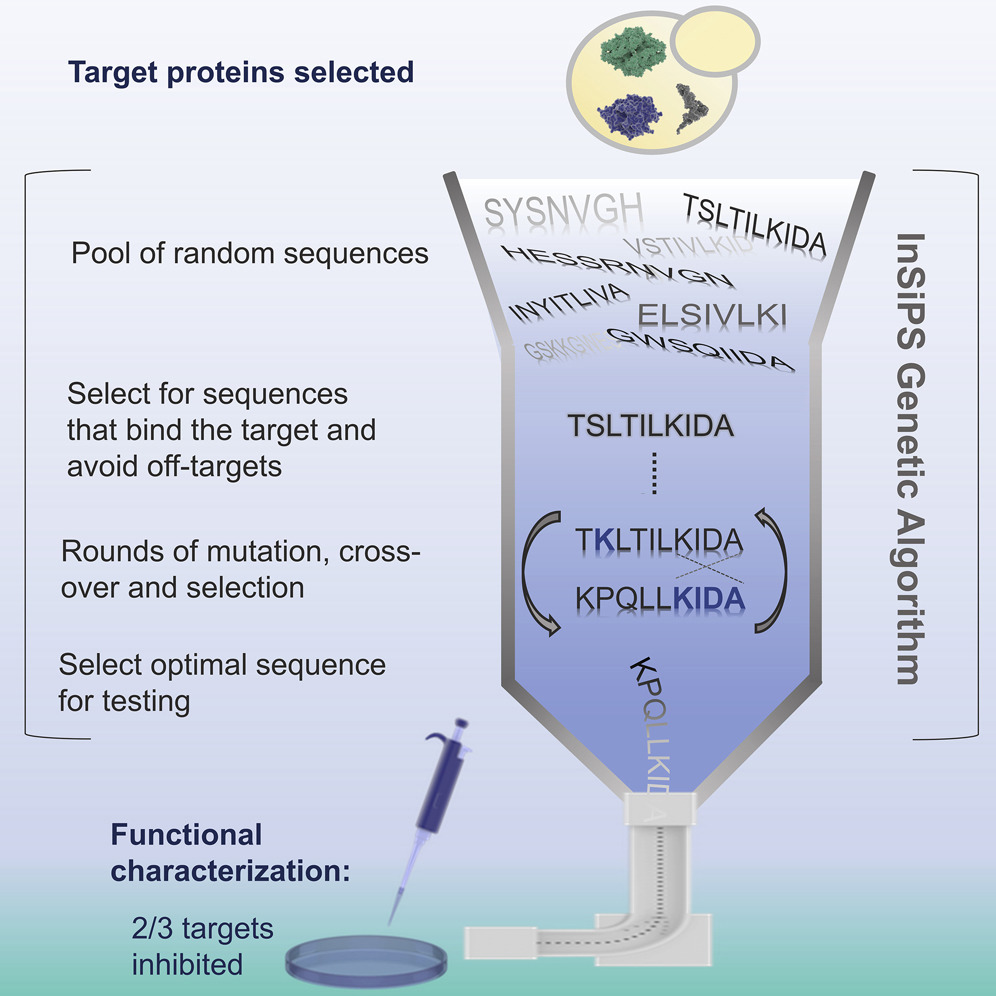
- In silico engineering of synthetic binding proteins from random amino acid sequences
Burnside, D., Schoenrock, A., Moteshareie, H., Hooshyar, M., Basra, P., Hajikarimlou, M., Dick, K., Barnes, B., Kazmirchuk, T., Jessulat, M., Pitre, S., Samanfar, B., Babu, M., Green, J.R., Wong, A., Dehne, F., Biggar, K.K., Golshani, A.
iScience 2019;11:375-387.
PMID:30660105
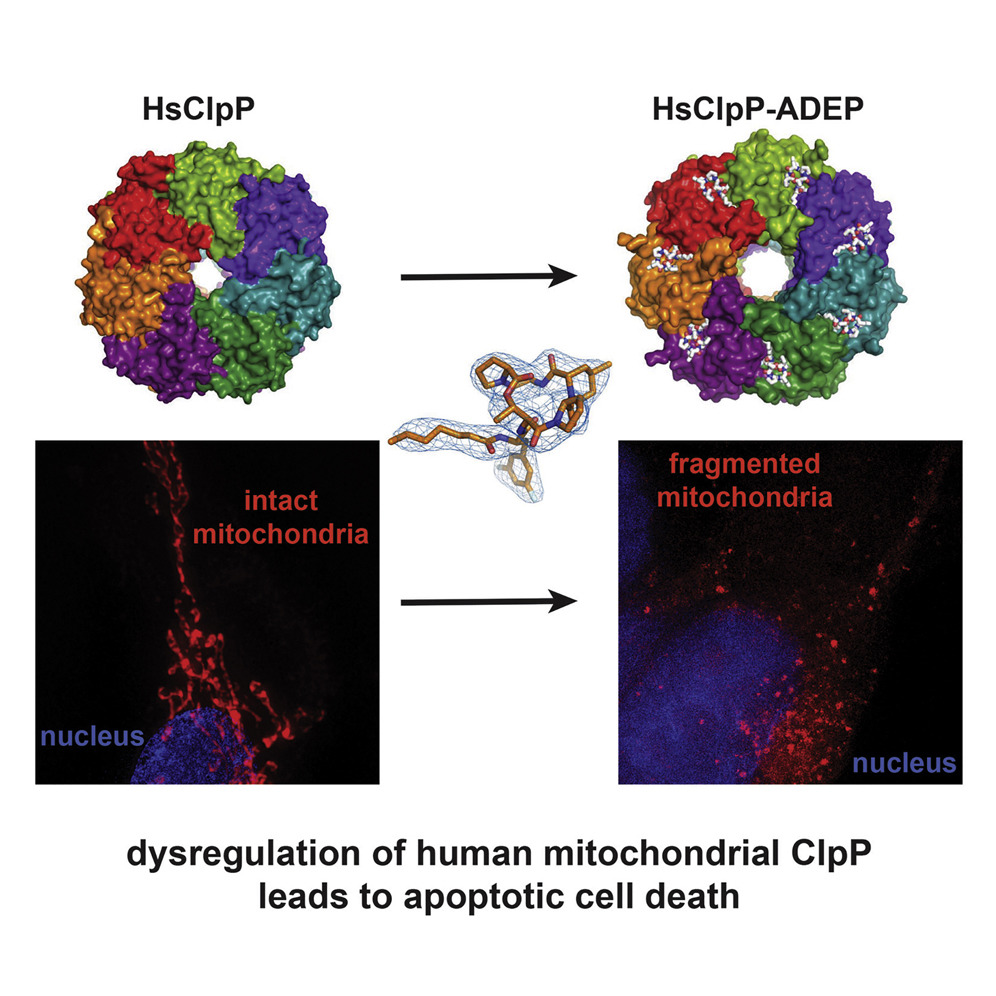
- Acyldepsipeptide Analogs Dysregulate Human Mitochondrial ClpP Protease Activity and Cause Apoptotic Cell Death
Wong, K. S., Mabanglo, M.F., Seraphim, T.V., Mollica, A., Mao, Y., Rizzolo, K., Leung, E., Moutaoufik, M. T., Hoell, L., Phanse, S., Goodreid, J., Barbosa, L.R.S., Ramos, C.H.I., Babu, M., Mennella, V., Batey, R.A., Schimmer, A.D., Houry, W.A.
Cell Chemical Biology. 2018;25(8):1017-1030.e9.
PMID:30126533
Preview
- EPHB6 augments both development and drug sensitivity of triple-negative breast cancer tumours
Toosi, B.M., El Zawily, A., Truitt, L., Shannon, M., Allonby, O., Babu, M., DeCoteau, J., Mousseau, D., Ali, M., Freywald, T., Gall, A.. Vizeacoumar, F. S., Kirzinger, M. W., Geyer, C.R., Anderson, D.H., Kim, T., Welm, A.L., Siegel, P., Vizeacoumar, F.J., Kusalik, A., & Freywald, A.
Oncogene 2018;37(30):4073-4093.
PMID:29700392
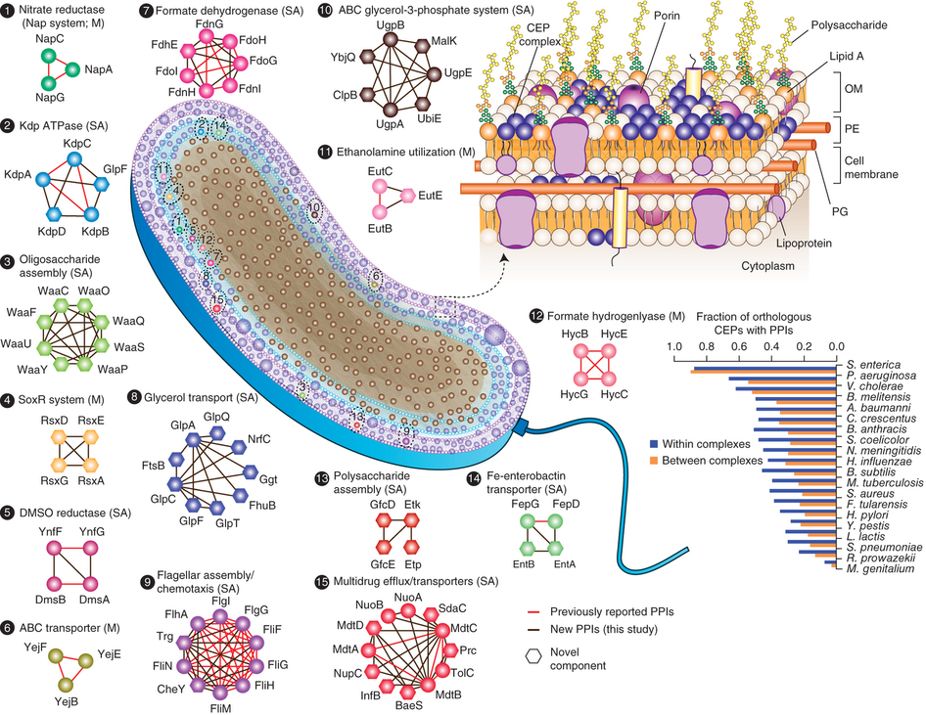
- Global landscape of cell envelope protein complexes in Escherichia coli
Babu, M.*, Bundalovic-Torma, C., Calmettes, C., Phanse, S., Zhang, Q., Jiang, Y., Minic, Z., Kim, S., Mehla, J., Gagarinova, A., Rodionova, I., Kumar, A., Guo, H., Kagan, O., Pogoutse, O., Aoki, H., Deineko, V., Caufield, J. H., Holtzapple, E., Zhang, Z., Vastermark, A., Pandya, Y., Lai, C. C., El Bakkouri, M., Hooda, Y., Shah, M., Burnside, D., Hooshyar, M., Vlasblom, J., Rajagopala, S.V., Golshani, A., Wuchty, S., Greenblatt, J.F., Saier, M., Uetz, P., F Moraes, T., Parkinson, J., Emili, A.
Nature Biotechnology 2018;36(1):103-112. (*corresponding)
PMID:29176613
Preview
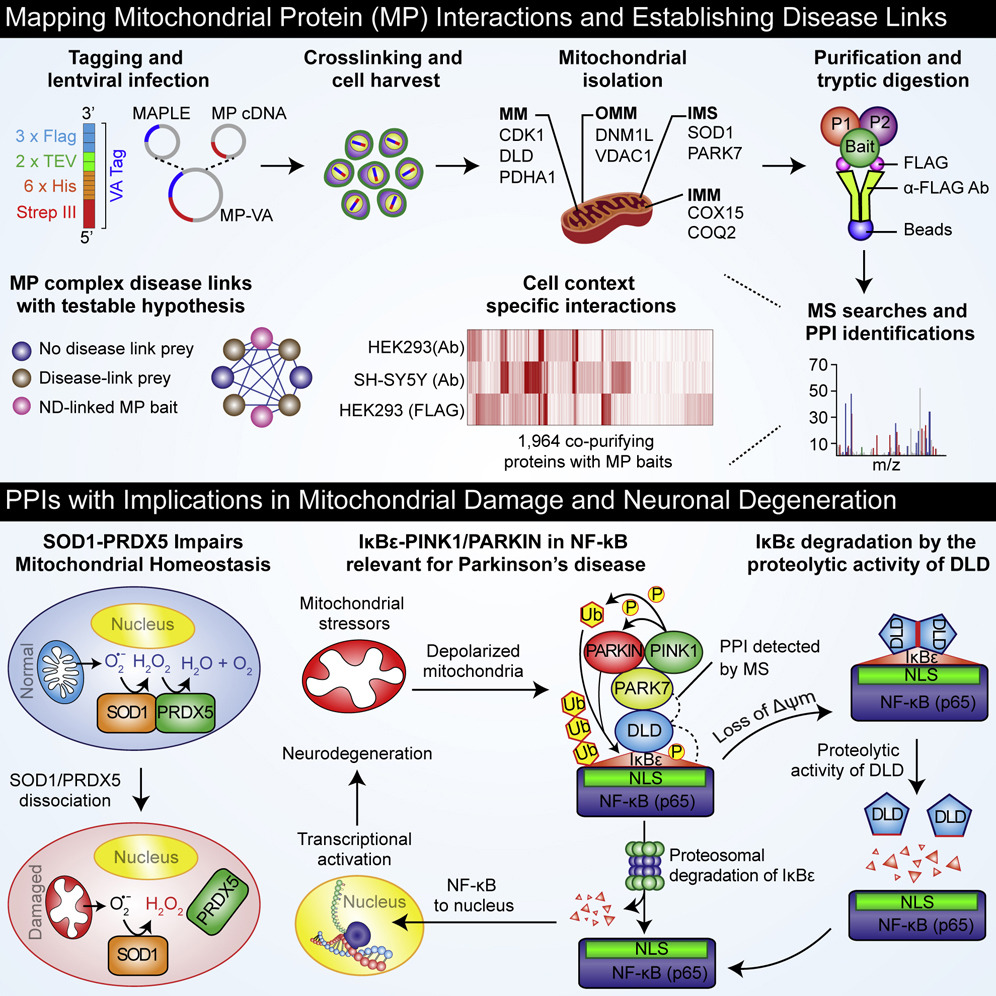
- A map of human mitochondrial protein interactions linked to neurodegeneration reveals new mechanisms of redox homeostasis and NF-κB signaling
Malty, R. H., Aoki, H., A., K., Phanse, S., Amin, S., Zhang, Q., Minic, Z., Goebels, F., Musso, G., Wu, Z., Abou-tok, H., Meyer, M., V., D., Kassir, S., Sidhu, V., Jessulat, M., Scott, N. E., Xiong, X., Vlasblom, J., Prasad, B., Foster, L. J., Alberio, T., Garavaglia, B., Yu, H., Bader, G. D., Nakamura, K., Parkinson, J., and Babu, M*.
Cell Systems 2017;5(6):564-577.e12. (*corresponding)
PMID:29128334
Preview
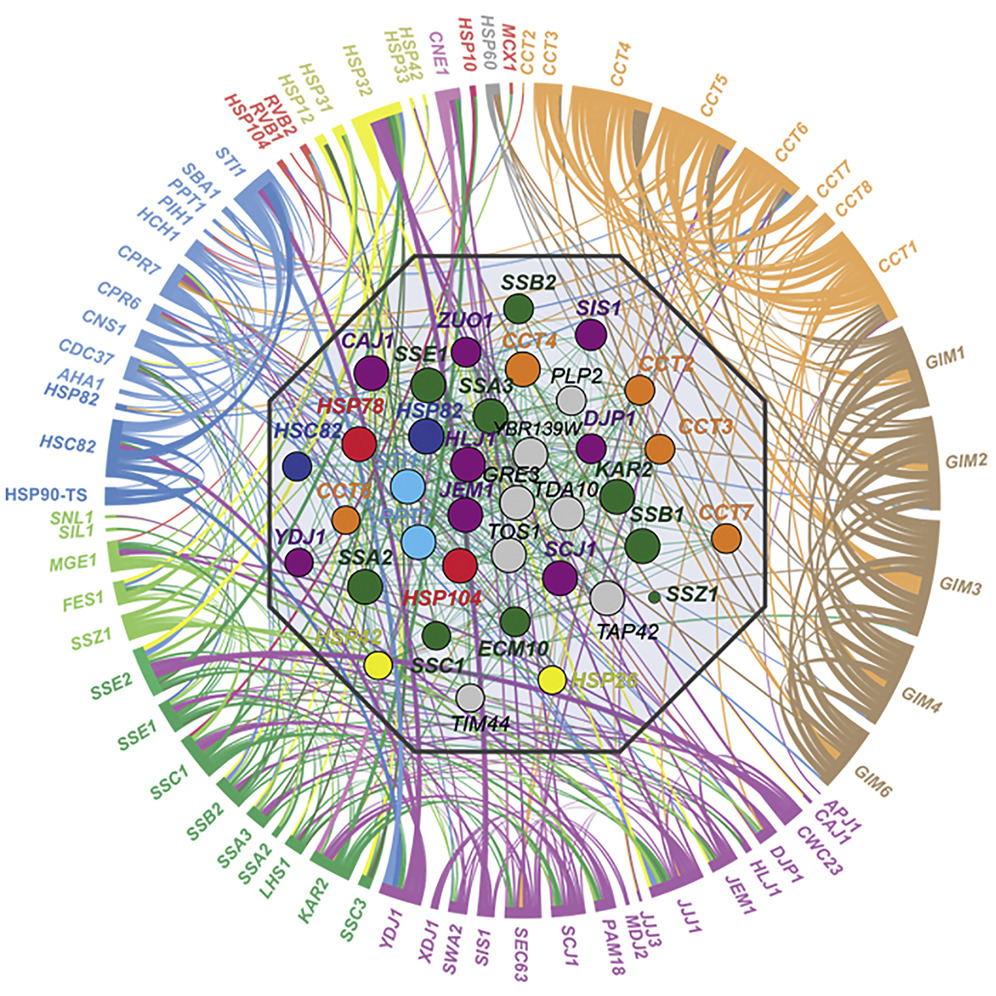
- Features of the Chaperone Cellular Network Revealed through Systematic Interaction Mapping
Rizzolo, K., Huen, J., Kumar, A., Phanse, S., Vlasblom, J., Kakihara,Y., Zeineddine, H. A., Minic, Z., Snider, J., Wang, W., Pons, C., Seraphim, T. V., Boczek, E. E., Alberti, S., Costanzo, M., Myers, C. L., Stagljar, I., Boone, C., Babu*, M., Houry*, W. A.
Cell Reports 2017;20(11):2735-2748. (*co-corresponding)
PMID:28903051
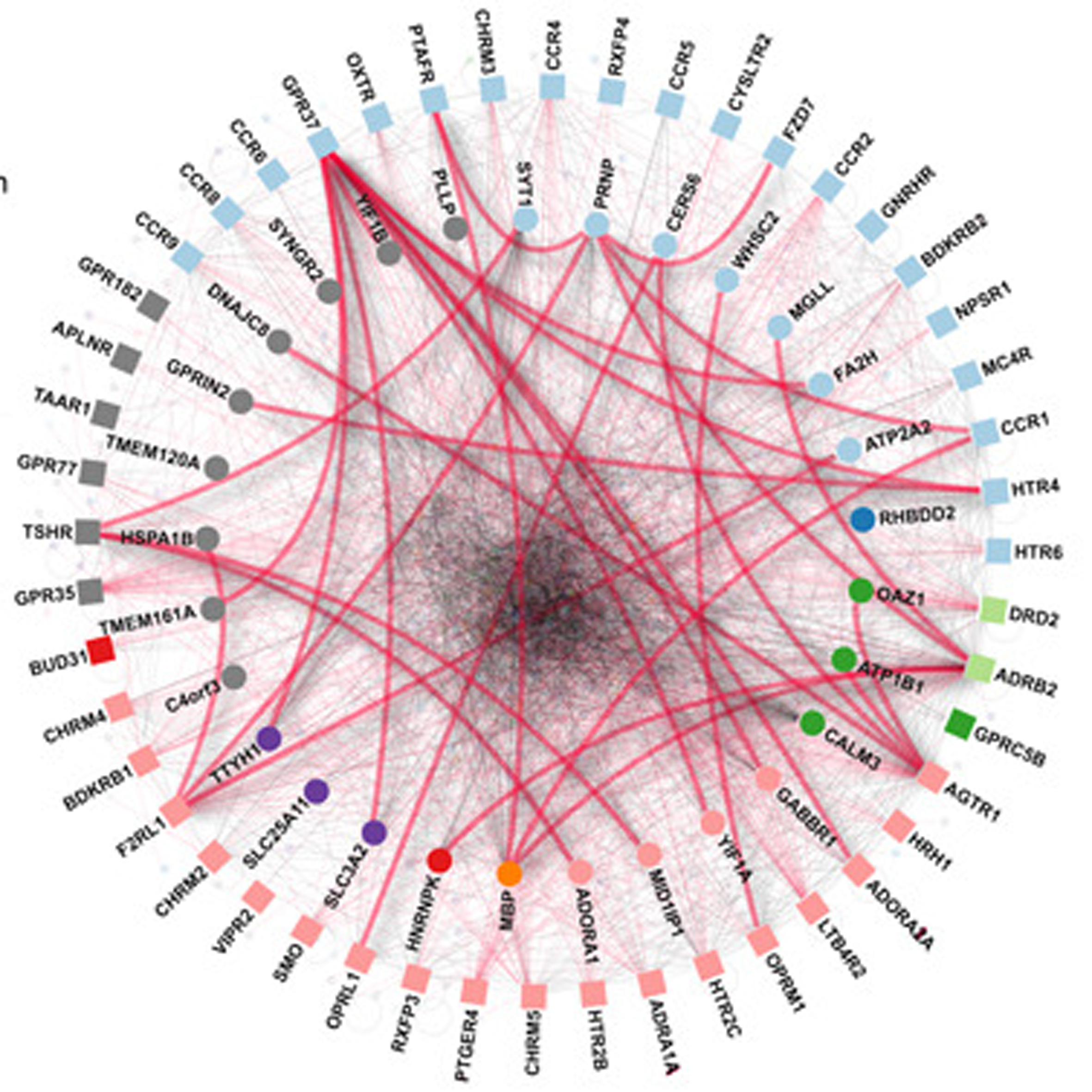
- Systematic protein–protein interaction mapping for clinically relevant human GPCRs
Sokolina, K., Kittanakom, S., Snider, J., Kotlyar, M., Maurice, P., Gandía, J., Benleulmi-Chaachoua, A., Tadagaki, K., Oishi, A., Wong, V., Malty, R.H., Deineko, V., Aoki, H., Amin, S., Yao, Z., Morató, X., Otasek, D., Kobayashi, H., Menendez, J., Auerbach, D., Angers, S., Pržulj, N., Bouvier, M., Babu, M., Ciruela, F., Jockers, R., Jurisica, I., Stagljar, I.
Molecular Systems Biology 2017;13(3):918.
PMID:28298427
- A Global Analysis of the Receptor Tyrosine Kinase-Protein Phosphatase Interactome
Yao, Z., Darowski, K., St-Denis, N., Wong, V., Offensperger, F., Villedieu, A., Amin, S., Malty, R., Aoki, H., Guo, H., Xu, Y., Iorio, C., Kotlyar, M., Emili, A., Jurisica, I., Neel, B.G., Babu, M., Gingras, A., Stagljar I.
Molecular Cell 2017;65(2):347-360.
PMID:28065597
- Systematic Genetic Screens Reveal the Dynamic Global Functional Organization of the Bacterial Translation Machinery
Gagarinova, A., Stewart, G., Samanfar, B., Phanse, S., White, C.A., Aoki, H., Deineko, V., Beloglazova, N., Yakunin, A.F., Golshani, A., Brown, E.D., Babu, M., Emili, A.
Cell Reports 2016;17(3):904-916.
PMID:27732863
- Conditional Epistatic Interaction Maps Reveal Global Functional Rewiring of Genome Integrity Pathways in Escherichia coli
Kumar, A., Beloglazova, N., Bundalovic-Torma, C., Phanse, S., Deineko, V., Gagarinova, A., Musso, G., Vlasblom, J., Lemak, S., Hooshyar, M., Minic, Z., Wagih, O., Mosca, R., Aloy, P., Golshani, A., Parkinson, J., Emili, A., Yakunin A.E., and Babu M.*
Cell Reports 2016;14(3):648-61. (*corresponding)
PMID:26774489
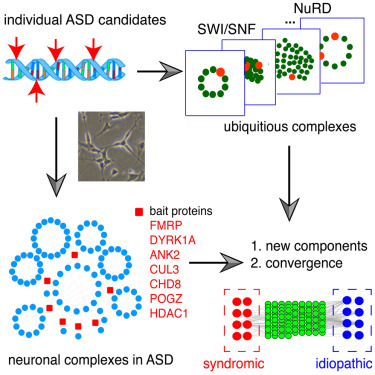
- Identification of Human Neuronal Protein Complexes Reveals Biochemical Activities and Convergent Mechanisms of Action in Autism Spectrum Disorders
Li, J., Ma, Z., Shi, M., Malty, R.H., Aoki, H., Minic, Z., Phanse, S., Jin, K., Wall, D.P., Zhang, Z., Urban, A.E., Hallmayer, J., Babu, M*., Snyder, M*.
Cell Systems 2015;1(5):361-374. (*co-corresponding)
PMID:26949739
Preview
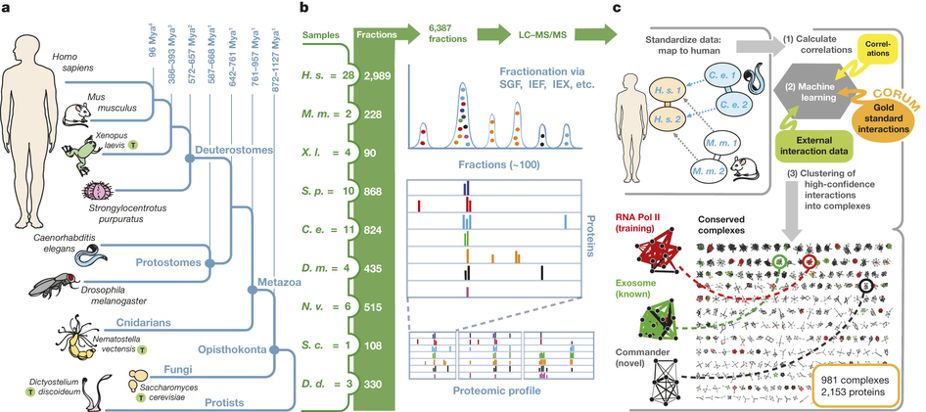
- Panorama of ancient metazoan macromolecular complexes
Wan, C., Borgeson, B., Phanse, S., Tu, F., Drew, K., Clark, G., Xiong, X., Kagan, O., Kwan, J.,Bezginov, A., Chessman, K., Pal, S., Cromar, G., Papoulas, O., Ni, Z., Boutz, D. R., Stoilova, S., Havugimana, P. C., Guo, X., Malty, R. H., Sarov, M., Greenblatt, J., Babu, M., Derry, W. B., Tillier, E. R., Wallingford, J. B., Parkinson, J., Marcotte, E. M. & Emili, A.
Nature 2015;525(7569):339-44.
PMID: 26344197
Preview
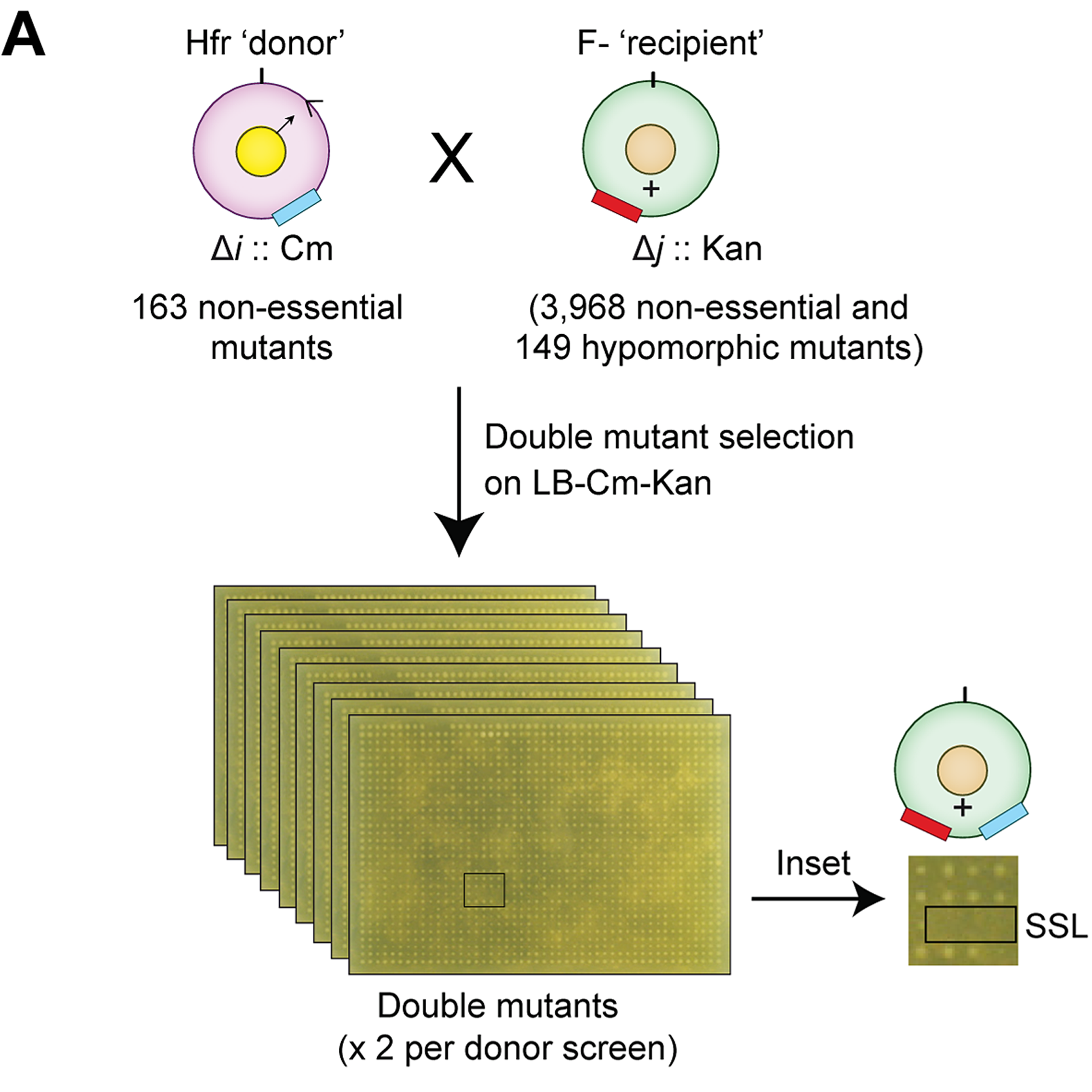
- Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli
Babu, M*, Arnold, A., Bundalovic-Torma, C., Gagarinova, A., Wong, K.S., Kumar, A., Stewart, G., Samanfar, B., Aoki, H., Wagih, O., Vlasblom, J., Phanse, S., Lad, K., Yu, AYH, Graham, C., Jin, K., Brown, E., Golshani, A., Kim, P., Moreno-Hagelsieb, G., Greenblatt, J., Houry, W.A., Parkinson, J. and Emili, A.
PLoS Genetics 2014;10(2):e1004120. (*corresponding)
PMID: 24586182
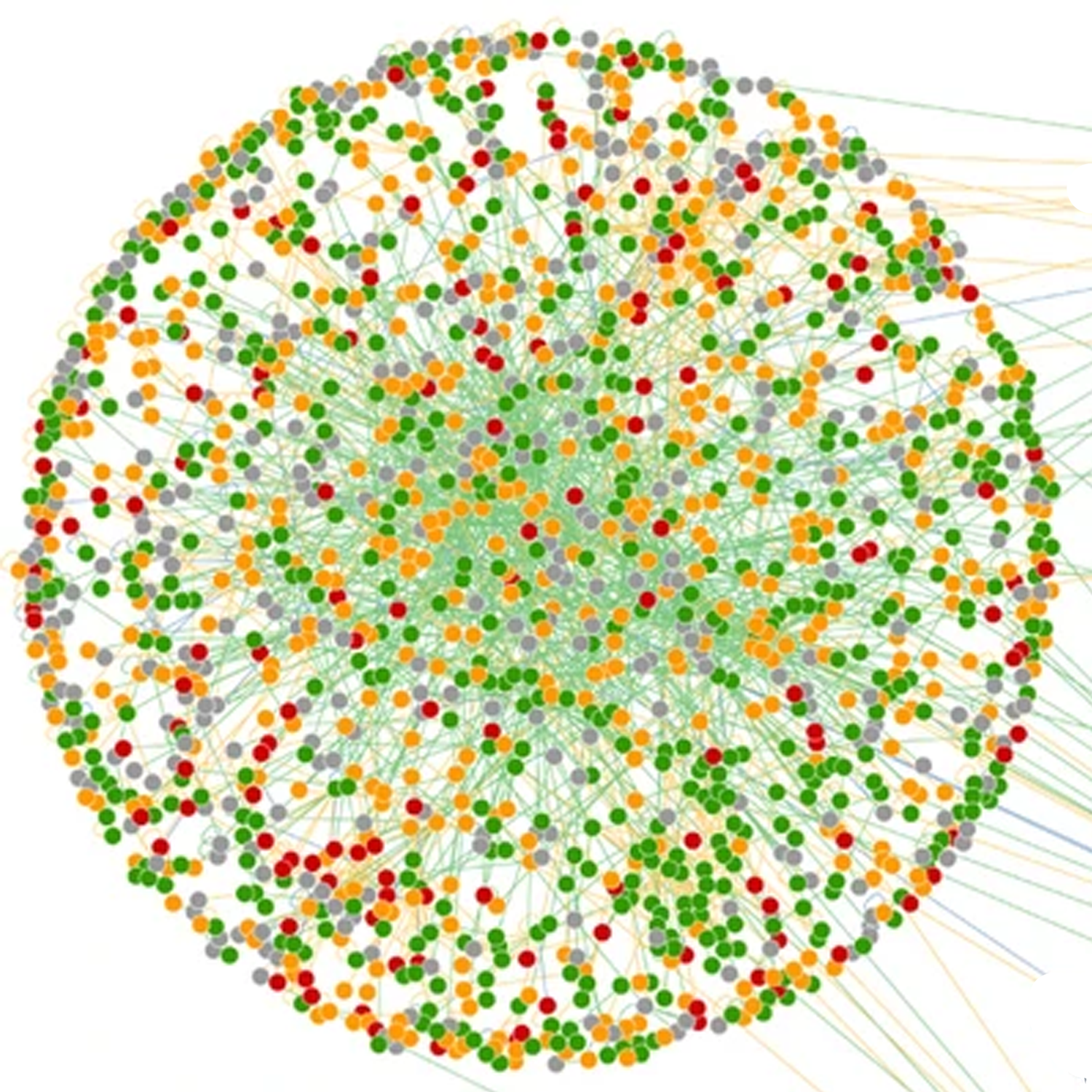
- Binary Protein-protein Interaction Landscape of Escherichia coli
Rajagopala, S.V*., Sikorski, P., Kumar, A., Mosca, R., Vlasblom, J., Arnold, R., Franca-Koh, J., Pakala, S.B., Phanse, S., Ceol, A., Häuser, R., Siszler, G., Wuchty, S., Emili, A., Babu, M*., Aloy, P., Pieper, R. and Uetz, P.
Nature Biotechnology 2014;32(3):285-290. (*co-corresponding)
PMID: 24561554
Preview
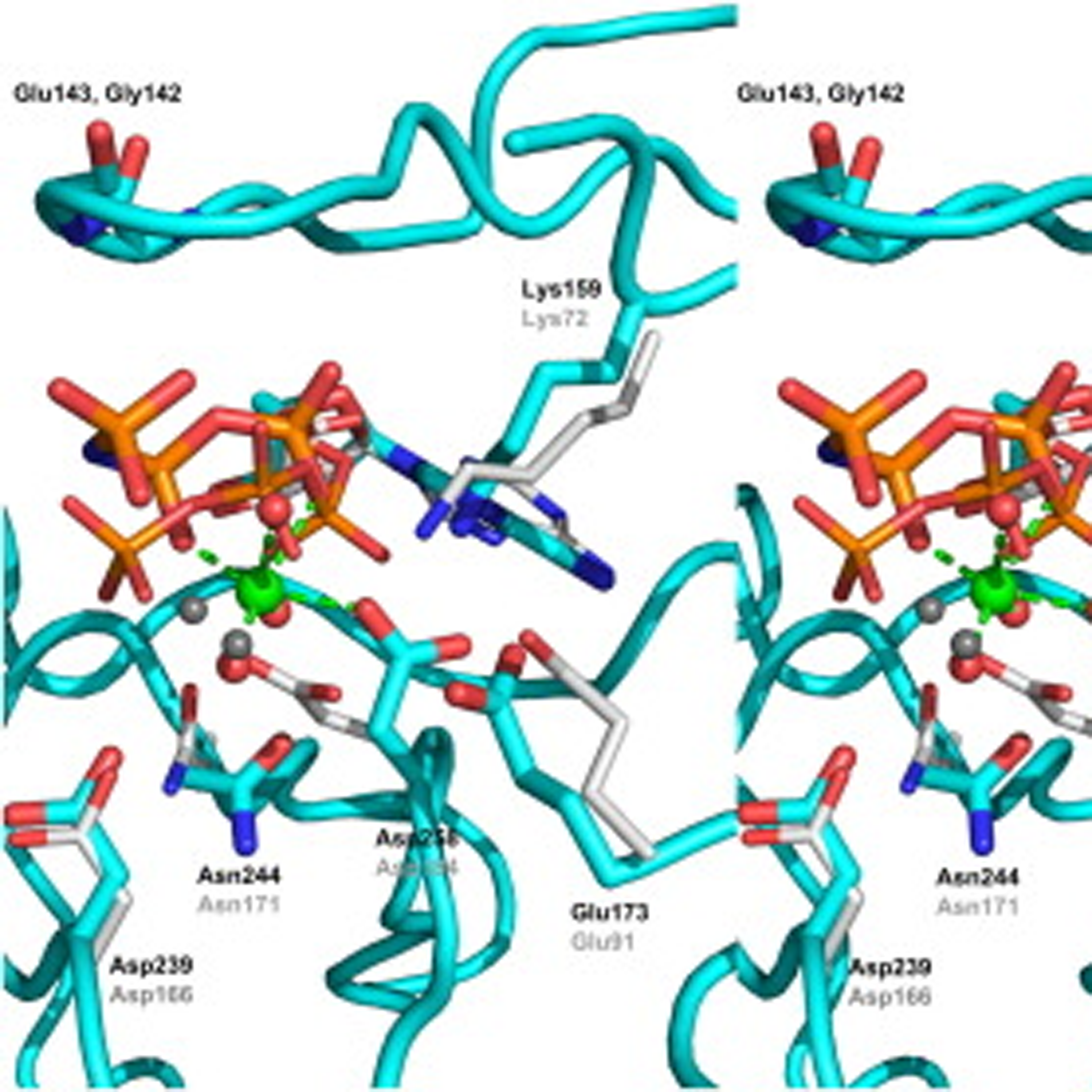
- NleH defines a new family of bacterial effector kinases
Grishin, A.M., Cherney, M., Anderson, D.H., Phanse, S., Babu, M. and Cygler, M.
Structure 2014;22(2):250-9.
PMID: 24373767
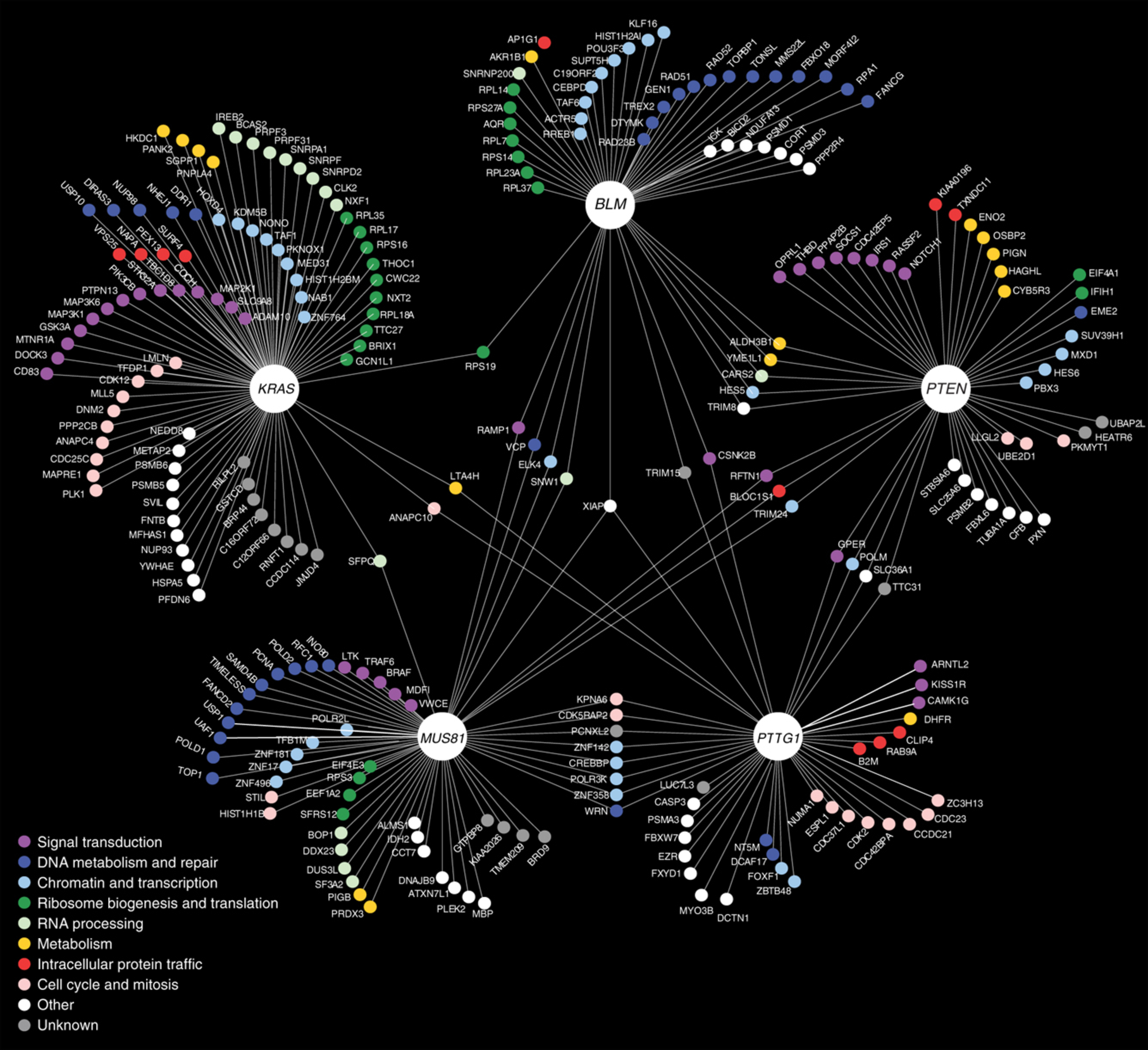
- A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities
Vizeacoumar, F. J., Arnold, R., Vizeacoumar, F. S., Chandrashekhar, M., Buzina, A., Young, J. T. F., Kwan, J.H.M., Sayad, A., Mero, P., Lawo, S., Tanaka, H., Brown, K. R., Baryshnikova, A., Mak, A. B., Fedyshyn, Y., Wang, Y., Brito, G. C., Kasimer, D., Makhnevych, T., Ketela, T., Datti, A., Babu, M., Emili, A., Pelletier L, Wrana J, Wainberg Z, Kim P. M., Rottapel R, O'Brien C. A., Andrews B, Boone C, Moffat J.
Molecular Systems Biology 2013;9:696.
PMID: 24104479
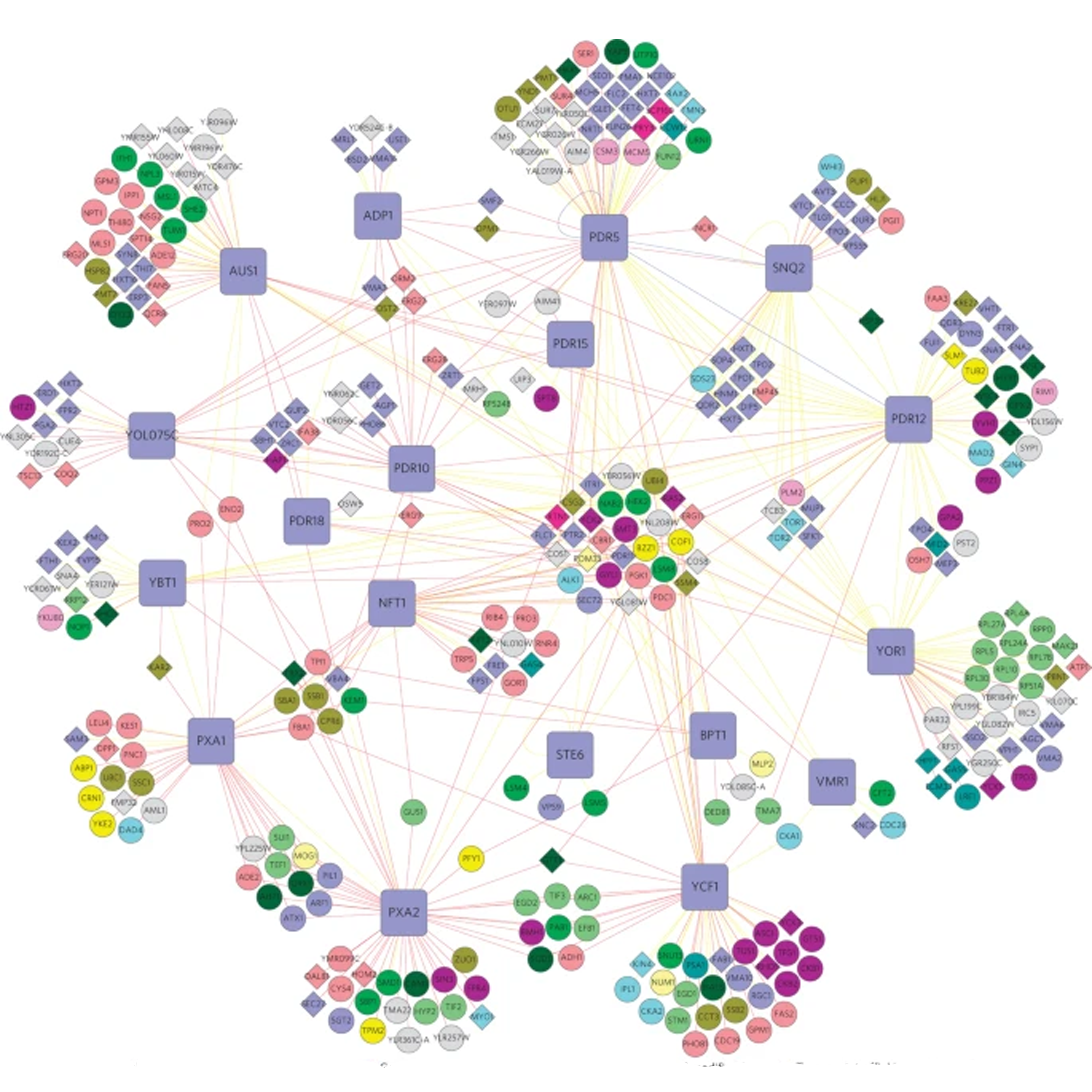
- Mapping the functional yeast ABC transporter interactome
Snider, J., Hanif, A. Lee, M. E., Jin, K., Yu, Q. R., Graham, C., Chuk, M., Damjanovic, D., Wierzbicka, M., Tang, P., Balderes, D., Wong, V., Jessulat, M., Darowski, K. D., San Luis, B., Shevelev, I., Sturley, S. L., Boone, C., Greenblatt, J. F., Zhang, Z., Paumi, C. M., Babu, M., Park, H., Michaelis S., & Stagljar, I.
Nature Chemical Biology 2013;9(9):565-72.
PMID: 23831759
Preview
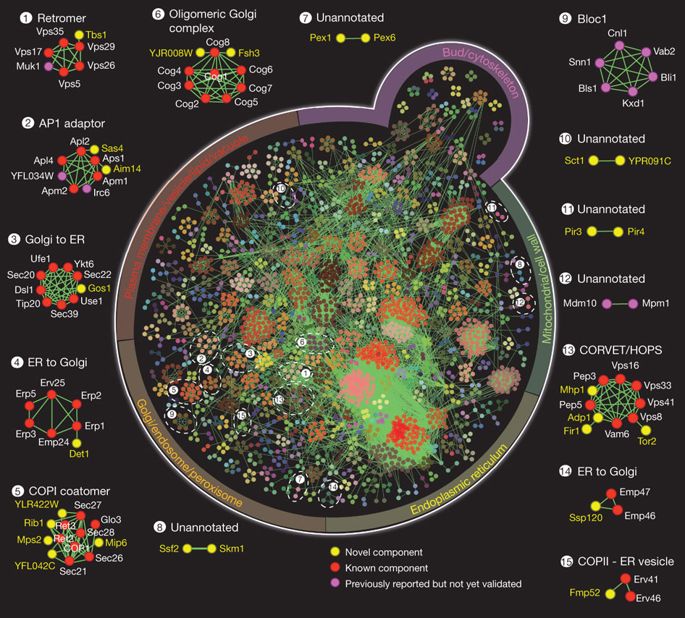
- Interaction landscape of membrane protein complexes in Saccharomyces Cerevisiae
Babu, M., Vlasblom, J., Pu, S., Guo, X., Graham, C., Vizeacoumar, F.J., Bean, B.D.M., Phanse, S., Fong, V., Davey, M., Snider, J., Hnatshak, O., Bajaj, N., Chandran, S., Punna, T., Christopolous, C., Wong, V., Zhong, G., Li, J., Stagljar, I., Conibear, E., Wodak, S.J.W., Emili, A. and Greenblatt, J.F.
Nature 2012;489:585-9.
PMID: 22940862
Preview
-
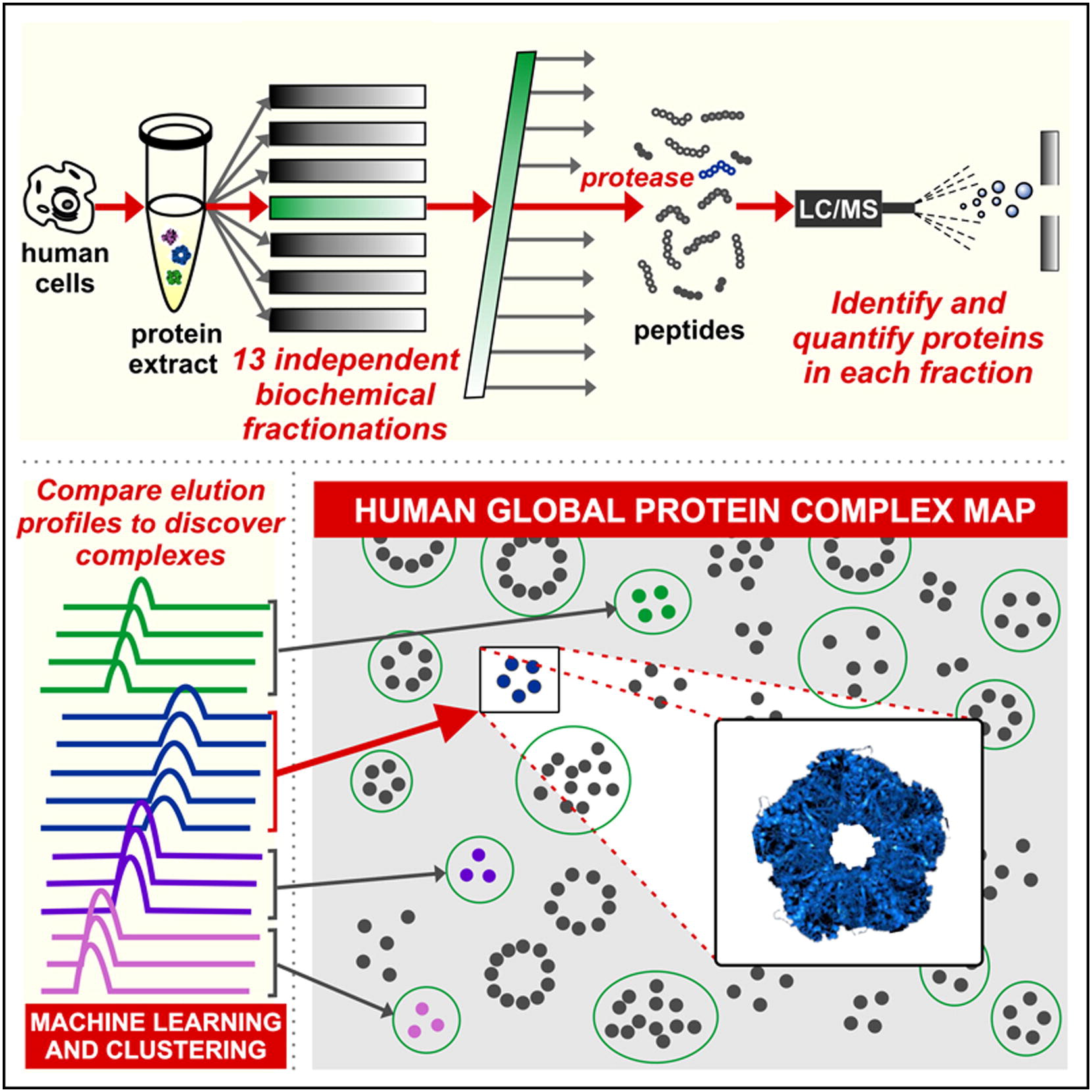
- A census of human soluble protein complexes
Havugimana, P.C., Hart, G.T., Nepusz, T., Yang, H., Turinsky, A.L., Li, Z., Wang, P.I., Boutz, D.R., Fong, V., Phanse, S., Babu, M., Craig, S.A., Hu, P., Wan, C., Vlasblom, J., Dar, V., Bezginov, A., Clark, G.W., Wu, G.C., Wodak, S.J., Tillier, E.R.M., Paccanaro, A., Marcotte, E.M., Emili, A.
Cell 2012;150:1068-1081.
PMID: 22939629
Preview
-
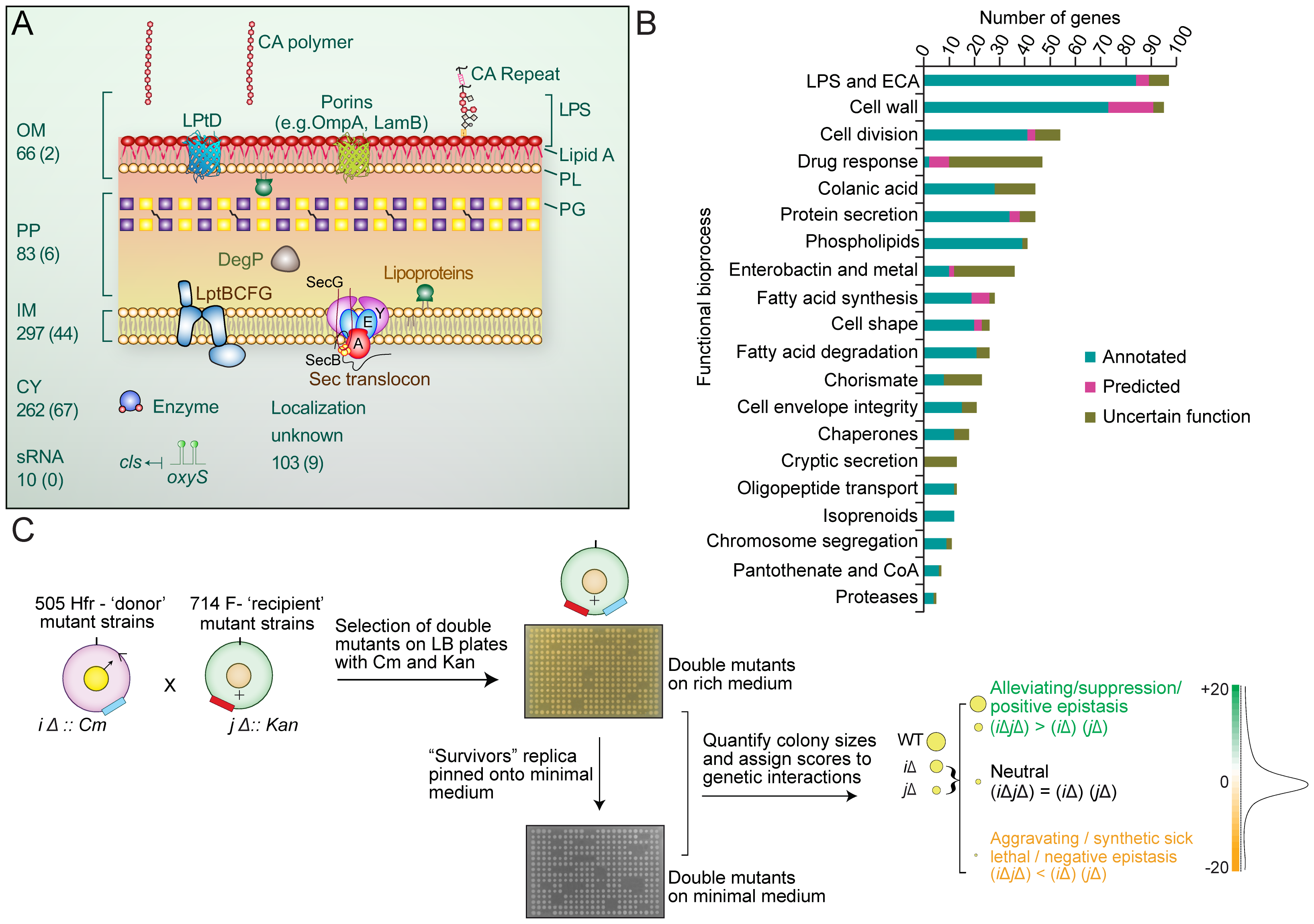
- Genetic interaction maps in Escherichia coli reveal condition-dependent functional crosstalk among cell envelope biogenesis pathways
Babu, M*., Díaz-Mejía, J.J., Vlasblom, J., Gagarinova, A., Phanse, S., Graham, C., Arnold, R., Yousif, F., Ding, H., Xiong, X., Nazarians-Armavil, A., Alamgir, Md., Ali, M., Pogoutse, O., Pe′er, A., Parkinson, J., Golshani, A., Whitfield, C., Wodak, S.J., Moreno-Hagelsieb, G., Greenblatt, J.F. and Emili, A.*
PLoS Genetics 2011;7(11):e1002377. (*co-corresponding)
PMID: 22125496
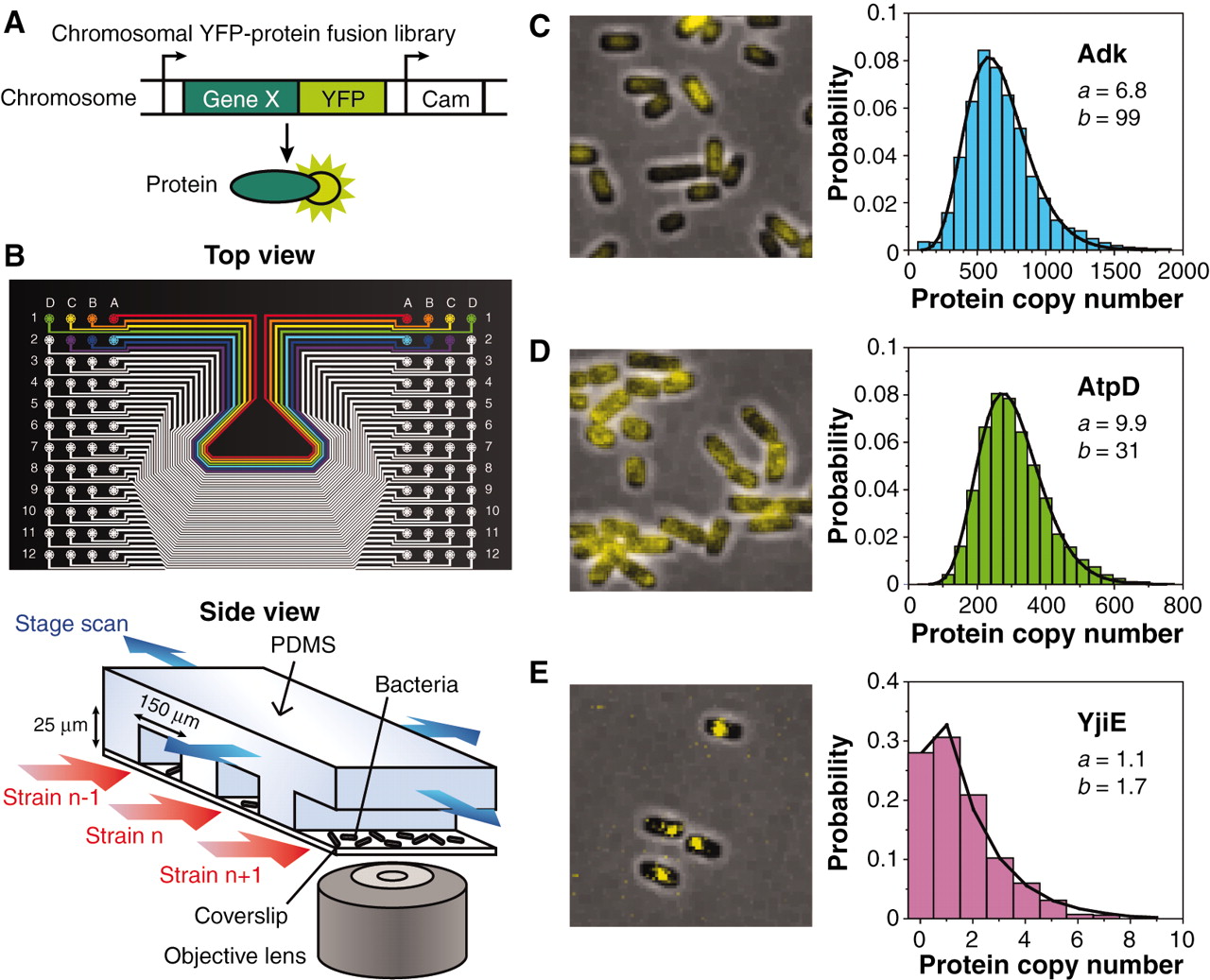
- Quantifying the E. coli proteome and transcriptome in a single cell with single-molecule sensitivity
Taniguchi, Y., Choi, P.J., Li, G-W., Chen, H., Babu, M., Hearn, J., Emili, A. and Xie, X.S.
Science 2010;329:533-538.
PMID: 20671182
Preview
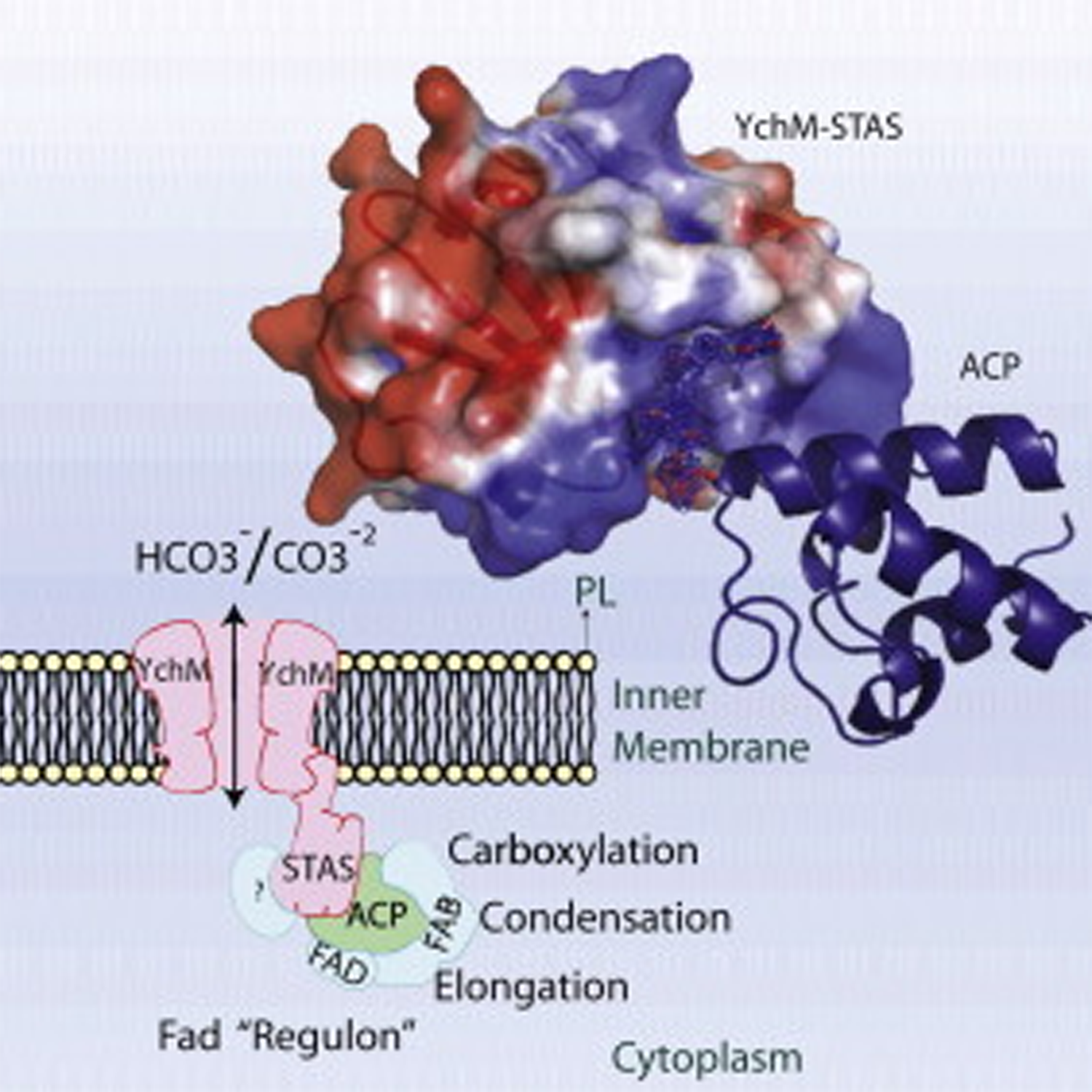
- Structure of a SLC26 anion transporter STAS domain in complex with acyl carrier protein: implications for E. coli YchM in fatty acid metabolism
Babu, M., Greenblatt, J., Emili, A., Strynadka, N. C.J., Reithmeier, A.F. and Moraes, T.F.
Structure 2010;18,:1450-1462.
PMID: 21070944
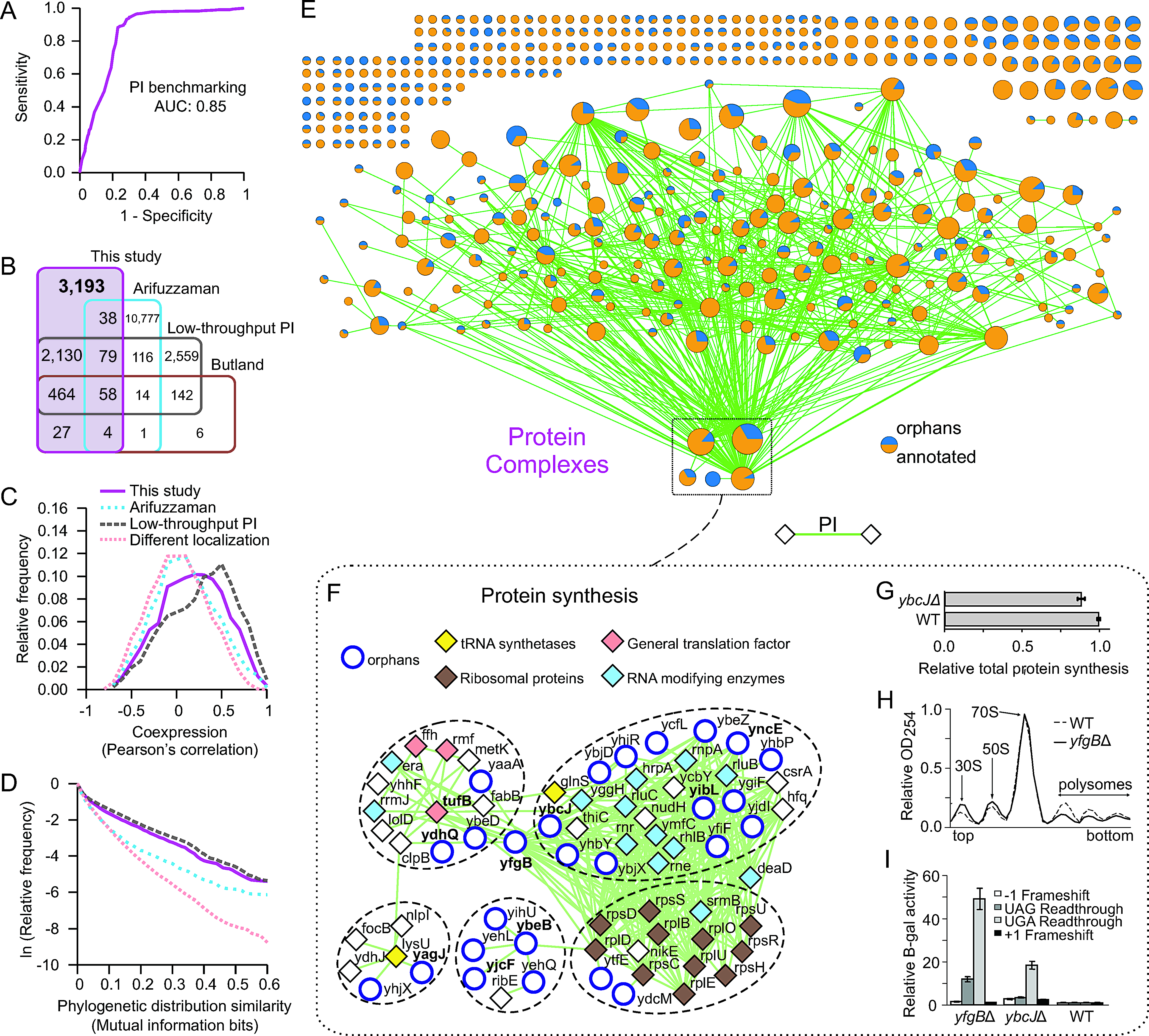
- Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins
Hu, P.*, Janga, S.C.*, Babu, M.*, Diaz-Mejia, J.J.*, Butland, G.*, Yang, W., Pogoutse, O., Guo, X., Phanse, S., Wong, P., Chandran, S., Christopoulos, C., Nazarians-Armavi,l A., Nasseri, N. K., Musso, G., Ali, M., Nazemof, N., Eroukova, V., Golshani, A., Paccanaro, A., Greenblatt, J. F., Moreno-Hagelsieb, G., Emili, A.
PLoS Biology 2009;7(4):e96. (*co-first author)
PMID: 19402753
Preview
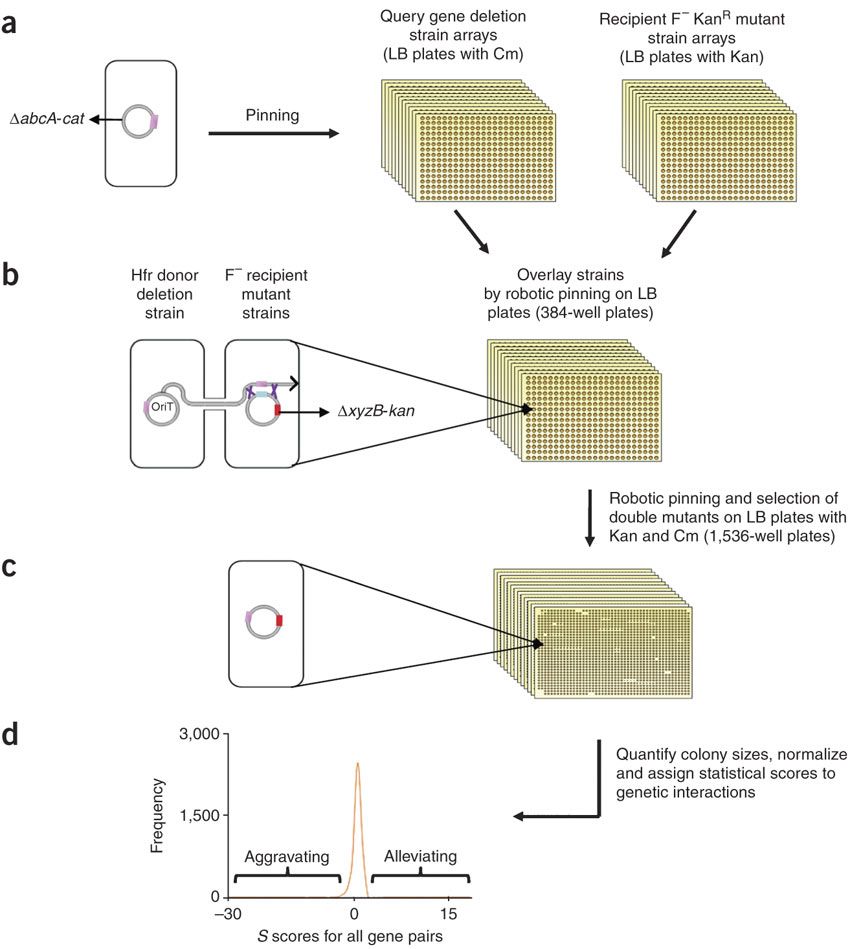
- eSGA: E. coli synthetic genetic array analysis
Butland, G.*, Babu, M.*, Diaz-Mejia, J.J., Bohdana, F., Phanse, S., Gold, B., Yang, W., Li, J., Gagarinova, A. G., Pogoutse, O., Mori, H., Wanner, B. L., Lo, H., Wasniewski, J., Christopolous, C., Ali, M., Venn, P., Safavi-Naini, A., Sourour, N., Caron, S., Choi, J. Y., Laigle, L., Nazarians-Armavil, A., Deshpande, A., Joe, S., Datsenko, K. A., Yamamoto, N., Andrews, B. J., Boone, C., Ding, H., Sheikh, B., Moreno-Hagelseib, G., Greenblatt, J. F., Emili, A.
Nature Methods 2008;5:89-795.(*co-first author)
PMID: 18677321
Preview
↑ Top
- Identification and Characterization of Protein Interactions with the Major Niemann-Pick Type C Disease Protein in Yeast Reveals Pathways of Therapeutic Potential
Hammond, N., JSnider, J., Stagljar, I., Mitchell, K., Lagutin, K., Jessulat, M., Babu, M., Teesdale-Spittle, P.H., Sheridan, J.P., Sturley, S.L., Munkacsi, A.B.
Genetics, iyad129, https://doi.org/10.1093/genetics/iyad129
PMID:37440478
- HDL functionality is dependent on hepatocyte stress defense factors Nrf1 and Nrf2
Trites M.J., Stebbings, B.M., Aoki, H., Phanse, S., Akl, M.G., Li, L., Babu, M., Widenmaier, S.B.
Frontiers in Physiology 2023 July 12;14. doi: 10.3389/fphys.2023.1212785.
PMID:37501930
- Sanitation enzymes: Exquisite surveillance of the noncanonical nucleotide pool to safeguard the genetic blueprint
Broderick, K., Moutaoufik, M.T., Aly, K.A., Babu, M.*
Seminars in Cancer Biology 2023 May 19;94:11-20. doi: 10.1016/j.semcancer.2023.05.005. Online ahead of print. (*corresponding)
PMID:37211293
- E. coli allantoinase is activated by the downstream metabolic enzyme, glycerate kinase, and stabilizes the putative allantoin transporter by direct binding
Rodionova, I.A., Hosseinnia, A., Kim, S., Goodacre, N., Zhang, L., Zhang, Z., Palsson, B., Uetz, P., Babu, M., & Saier Jr., M.H.
Scientific Reports 2023 May 5;13(1):7345. doi: 10.1038/s41598-023-31812-4.
PMID:37147430
- Complementary gene regulation by NRF1 and NRF2 protects against hepatic cholesterol overload
Akl, M.G., Li, L., Baccetto, R., Phanse, S., Zhang, Q., Trites, M.J., McDonald, S., Aoki, H., Babu, M., Widenmaier, S.B.
Cell Reports 2023 Apr 14;42(4):112399. doi: 10.1016/j.celrep.2023.112399
PMID:337060561
- A Correlation between 3′-UTR of OXA1 Gene and Yeast Mitochondrial Translation
Hajikarimlou, M., Hooshyar, M., Sunba, N., Nazemof, N., Moutaoufik, M.T., Phanse, S., Said, K.B., Babu, M., Holcik, M., Samanfar, B., Smith, M., and Golshani, A.
Journal of Fungi 2023, Apr 5;9(4), 445; https://doi.org/10.3390/jof9040445
↑ Top
- Insights into SACS pathological attributes in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS)☆
Aly, K.A., Moutaoufik, M.T., Zilocchi, M., Phanse, S., Babu, M.*
Current Opinion Chemical Biology. 2022 Sep 17;71:102211. (*corresponding)
PMID:36126381
- A computational approach to rapidly design peptides that detect SARS-CoV-2 surface protein S
Hajikarimlou, H., Hooshyar, M., Moutaoufik, M.T., Aly, K.A., Azad, T., Takallou, S., Jagadeesan, S., Phanse, S., Said, K.B., Samanfar, B., Bell, J.C., Dehne, F., Babu, M., Golshani, A.
NAR Genomics and Bioinformatics, Volume 4, Issue 3, September 2022, lqac058 (*corresponding)
PMID:36004308
- In vitro toxicity screening of amorphous silica nanoparticles using mitochondrial fraction exposure followed by MS-based proteomic analysis.
Kumarathasan, P., Nazemof,N.,Breznan, D., Blais, E., Aoki, H., Gomes, J., Vincent, R., Phanse, S. and Babu M.
Analyst. 2022 Aug 8;147(16):3692-3708.
PMID:35848500
- Auxotrophic and prototrophic conditional genetic networks reveal the rewiring of transcription factors in Escherichia coli.
Gagarinova, A., Hosseinnia, A., Rahmatbakhsh, M., Istace, Z., Phanse, S., Moutaoufik, M.T., Zilocchi, M., Zhang, Q., Aoki, H., Jessulat, M., Kim, S., Aly, K.A., & Babu, M.*
Nature Communications 2022 Jul 14;13(1):4085 (*corresponding)
PMID:35835781
- Development of a Method Combining Peptidiscs and Proteomics to Identify, Stabilize, and Purify a Detergent-Sensitive Membrane Protein Assembly
Young, J.W., Wason, I.S., Zhao, Z., Kim, S., Aoki, H., Phanse, S., Rattray, D.G., Foster, L.J., Babu, M., Duong van Hoa, F. 1
Journal Proteome Research, 2022 Jul 1;21(7):1748-1758
PMID:35616533
- HPiP: an R/Bioconductor package for predicting host–pathogen protein–protein interactions from protein sequences using ensemble machine learning approach
Rahmatbakhsh, M., Moutaoufik, M.T., Gagarinova, G., Babu, M.*
Bioinformatics Advances, 2022 May 23;2(1):vbac038. (*corresponding)
PMID:35669347
- CFTR interactome mapping using the mammalian membrane two-hybrid high-throughput screening system
Lim, S.H., Snider, J., Birimberg-Schwartz, L., Ip, W., Serralha, J.C., Botelho, H.M., Lopes-Pacheco, M., Pinto, M.C, Moutaoufik, M.T., Zilocchi, M., Laselva, O., Esmaeili, M., Kotlyar, M., Lyakisheva, A., Tang, P., Vázquez, L.L., Akula, I., Aboualizadeh, F., Wong, V., Grozavu, I., Opacak-Bernardi, T., Yao, Z., Mendoza. M., Babu, M., Jurisica, I., Gonska, T., Bear, C.E., Amaral, M.D., Stagljar, I.
Molecular Systems Biology 2022 Feb;18(2):e10629.
PMID:35156780
- Adenosine A1 receptor ligands bind to α-synuclein: implications for α-synuclein misfolding and α-synucleinopathy in Parkinson's disease
Jakova, E., Moutaoufik, M.T., Lee, J.S., Babu, M., Cayabyab.F.S.
Translational Neurodegenertion 2022 Feb 10;11(1):9.
PMID:35139916
- The Thiazole-5-Carboxamide GPS491 Inhibits HIV-1, Adenovirus, and Coronavirus Replication by Altering RNA Processing/Accumulation
Dahal, S., Cheng, R., Cheung, P.K., Been, T., Malty, R., Geng, M., Manianis, S., Shkreta, L., Jahanshahi, S., Toutant, J., Chan, R., Park, S., Brockman, M.A., Babu, M., Mubareka, S., Mossman, K., Banerjee, A., Gray-Owen, S., Brown, M., Houry, W.A., Chabot, B., Grierson, D., Cochrane, A.
Viruses 2021 Dec 30;14(1):60.
PMID:35062264
- Assembly principles of the human R2TP chaperone complex reveal the presence of R2T and R2P complexes
Seraphim, T.V., Nano, N., Cheung, Y.W.S., Aluksanasuwan, S., Colleti, C., Mao, Yu-Q., Bhandari, V., Young, G., Höll, L., Phanse, S., Gordiyenko, Y., Southworth, D.R., Robinson, C.V., Thongboonkerd, V., Gava, L.M., Borges, J.C., Babu, M., Barbosa, L.R.S., Ramos, C.H.I., Kukura, P., Houry, W.A.
Structure . 2022 Jan 6;30(1):156-171.e12.
PMID:34492227
↑ Top
- Quantitative Genetic Screens for Mapping Bacterial Pathways and Functional Networks
Gagarinova, A., Hosseinnia, A., Babu, M.
Methods Molecular Biology. 2021;2381:3-37
PMID:34590268
- Human-Soybean Allergies: Elucidation of the Seed Proteome and Comprehensive Protein-Protein Interaction Prediction
Dick, K., Pattang, A., Hooker, J., Nissan, N., Sadowski, M., Barnes, B., Tan, L,H., Burnside, D., Phanse, S., Aoki, H., Babu, M., Dehne, F., Golshani, A., Cober, E.L., Green, J.R., Samanfar, B.
Journal Proteome Research 2021 Nov 5;20(11):4925-4947.
PMID:34582199
- A Panoramic View of Proteomics and Multiomics in Precision Health
Zilocchi, M., Wang, C., Babu, M.*, Li, J.*
iScience, 2021 Jul 30;24(8) (*co-corresponding)
- Actin-Related Protein 6 (Arp6) Influences Double-Strand Break Repair in Yeast
Hooshyar, M., Burnside, D., Hajikarimlou, M., Omidi, K., Jesso, A., Vanstone, M., Young, A.,Cherubini, P.M., Jessulat, M., Potter, T., Schoenrock, A., Bhojoo, U., Silva, E., Moteshareie, H., Babu, M., Diallo, J., Dehne, F., Samanfar, B., and Golshani, A.
Appl. Microbiol. 2021, 1(2), 225-238
- The conserved Tpk1 regulates non-homologous end joining double-strand break repair by phosphorylation of Nej1, a homolog of the human XLF
Jessulat, M., Amin, S., Hooshyar, M., Malty, R., Moutaoufik, M.T., Zilocchi, M., Istace, Z., Phanse, S., Aoki, H., Omidi, K., Burnside, D., Samanfar, B., Aly, K.A., Golshani, A., Babu, M.*
Nucleic Acids Research, 2021 Aug 20;49(14):8145-8160 (*corresponding)
PMID:34244791
- Bioinformatic Analysis of Temporal and Spatial Proteome Alternations During Infections
Rahmatbakhsh, M., Gagarinova, G.*, Babu, M.*
Frontiers in Genetics., 2021 Jul 2;12:667936. (*co-corresponding)
PMID:34276775
- ZapG (YhcB/DUF1043), a novel cell division protein in gamma-proteobacteria linking the Z-ring to septal peptidoglycan synthesis
Mehla, J., Liechti, G., Morgenstein, R.M., Caufield, J.H., Hosseinnia, A., Gagarinova, A., Phanse, S., Goodacre, N., Brockett, M., Sakhawalkar, N., Babu, M., Xiao, R., Montelione, G.T., Vorobiev, S., Blaauwen, T. den, Hunt, J.F., Uetz, P.
Journal of Biological Chemistry 2021;296:100700.
33895137
- Toward the discovery of biological functions associated with the mechanosensor Mtl1p of Saccharomyces cerevisiae via integrative multi-OMICs analysis
Martínez-Matías, M., Chorna, N., González-Crespo, S., Villanueva, L., Montes-Rodríguez, I., Melendez-Aponte, L.M., Roche-Lima, A., Carrasquillo-Carrión, K., Santiago-Cartagena, E., Rymond, B.C., Babu, M., Stagljar I. & Rodríguez-Medina, J.R.
Scientific Reports 2021;11(1):7411.
PMID:33795741
- From Fuzziness to Precision Medicine: On the Rapidly Evolving Proteomics with Implications in Mitochondrial Connectivity to Rare Human Disease
Aly, K.A., Moutaoufik, M.T., Phanse, S., Zhang, Q., Babu M.*
iScience 2021;24(2). (*corresponding)
PMID:33521598
- Functional cooperativity between the trigger factor chaperone and the ClpXP proteolytic complex
Rizzolo, K., Yu, A. H. Y., Ologbenla, A., Kim, S., Zhu, H., Ishimori, K., Thibault, G., Leung, E., Zhang, Y.W., Teng, M., Haniszewski, M., Miah, N., Phanse, S., Minic, Z., Lee, S., Caballero, J.D., Babu, M., Tsai, F.T.F., Saio T., & Houry, W.A.
Nature Communications 2021;12(1):281.
PMID:33436616
↑ Top
- Deletion of yeast TPK1 reduces the efficiency of non-homologous end joining DNA repair
Hooshyar, M., Jessulat, M., Burnside, D., Kluew, A., Babu, M.*, Golshani, A.
Biochemical and Biophysical Research Communications 2020;533(4):899-904. (*co-corresponding)
PMID:33008596
- Mitochondria Under the Spotlight: On the Implications of Mitochondrial Dysfunction and its Connectivity to Neuropsychiatric Disorders
Zilocchi, M., Broderick, K., Phanse, S., Aly, K.A., Babu M.*
Computational and Structural Biotechnology Journal 2020;18:2535-2546. (*corresponding)
PMID:33033576
- Lithium Chloride Sensitivity in Yeast and Regulation of Translation
Hajikarimlou, M., Hunt, K., Kirby, G., Takallou, S., Jagadeesan, S.K., Omidi, K., Hooshyar, M., Burnside, D., Moteshareie, H., Babu, M., Smith, M., Holcik, M., Samanfar, B., Golshani, A.,
Int. J. Mol. Sci. 2020;21(16):5730.
PMID:32785068
- Sensitivity of yeast to lithium chloride connects the activity of YTA6 and YPR096C to translation of structured mRNAs
Hajikarimlou, M., Moteshareie, H., Omidi, K., Hooshyar, M., Shaikho, S., Kazmirchuk, T., Burnside, D., Takallou, S., Zare, N., Jagadeesan, S. K., Puchacz, N., Babu, M., Smith, M., Holcik, M., Samanfar, B., Golshani, A.,
PLOS One 2020 Jul 8;15(7):e0235033.
PMID:32639961
- Protein Interactions of the Mechanosensory Proteins Wsc2 and Wsc3 for Stress Resistance in Saccharomyces cerevisiae
Vélez-Segarra, V., González-Crespo, S., Santiago-Cartagena, E., Vázquez-Quiñones, L. E., Martínez-Matías, N., Otero, Y., Zayas, J. J., Siaca, R., del Rosario, J., Mejías, I., Aponte, J. A., Collazo, N. C., Lasso, F. J., Snider, J., Jessulat, M., Aoki, H., Rymond, B. C., Babu, M., Stagljar, I., and Rodríguez-Medina, J. R.
G3: GENES, GENOMES, GENETICS 2020;10(9):3121-3135.
PMID:32641451
- Exploring the impact of PARK2 mutations on the total and mitochondrial proteome of human skin fibroblasts
Zilocchi, M., Colugnat, I., Lualdi, M., Meduri, M., Marini, F., Carregari, V.C., Moutaoufik, M.T., Phanse, S., Pieroni, L., Babu, M., Garavaglia, B., Fasano, M. and Alberio, T.
Front. Cell Dev. Biol.2020;8:423.
PMID:32596240
- BraInMap Elucidates the Macromolecular Connectivity Landscape of Mammalian Brain
Pourhaghighi, R., Ash, P.E.A., Phanse, S., Goebels, F., Hu, L.Z.M., Chen, S., Zhang, Y., Wierbowski, S.D., Boudeau, S., Moutaoufik, M.T., Malty, R.H., Malolepsza, E., Tsafou, K., Nathan, A., Cromar, G., Guo, H., Abdullatif, A.A., Apicco, D.J., Becker, L.A., Gitler, A.D., Pulst, S.M., Youssef, A., Hekman, R., Havugimana, P.C., White, C.A., Blum, B.C., Ratti, A., Bryant, C.D., Parkinson, J., Lage, K., Babu, M., Yu, H., Bader, G.D., Wolozin, B., Emili, A.
Cell Systems, 2020;10(4):333-350.e14.
PMID:32325033
- HERG channel and cancer: A mechanistic review of carcinogenic processes and therapeutic potential
He, S., Moutaoufik, M.T., Islam, S., Persad, A., Wu, A., Aly, K.A., Fonge, H., Babu, M., Cayabyab, F.S.
Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 2020;1873(2).
PMID:32135169
- Misconnecting the dots: altered mitochondrial protein-protein interactions and their role in neurodegenerative disorders
Zilocchi, M., Moutaoufik, M.T., Jessulat, M., Phanse, S., Aly, K.A., & Babu, M.*
Expert Review of Proteomics, 2020 Feb;17(2):119-136. (*corresponding)
PMID:31986926
↑ Top
- Chemical-genetic profiling reveals limited cross-resistance between antimicrobial peptides with different modes of action
Kintses, B., Jangir, P.K., Fekete, G., Számel, M., Méhi, O., Spohn, R., Daruka, L., Martins, A., Hosseinnia, A., Gagarinova, A., Kim, S., Phanse, S., Csörgő, B., Györkei, A., Ari, E., Lázár, V., Nagy, I., Babu, M., Pál C., & Papp, B.
Nature Communications 2019;10(1):5731.
PMID:31844052
- ClpP Protease Activation Results from the Reorganization of the Electrostatic Interaction Networks at the Entrance Pores
Mabanglo, M.F., Leung, E., Vahidi, S., Seraphim, T.V., Eger, B.T., Bryson, S., Bhandari, V., Zhou, J.L., Mao, Y., Rizzolo, K., Barghash, M.M., Goodreid, J.D., Phanse, S., Babu, M., Barbosa, L.R.S., Ramos, C.H.I., Batey, R.A., Kay, L.E., Pai, E.F., Houry, W.A.
Communications Biology - Nature 2019;2:410.
PMID:31754640
- Rewiring of the Human Mitochondrial Interactome During Neuronal Reprogramming Reveals Regulators of the Respirasome and Neurogenesis
Moutaoufik, M.T., Malty, R., Amin, S., Zhang, Q., Phanse, S., Gagarinova, A., Zilocchi, M., Hoell, L., Minic, Z., Gagarinaova, M., Aoki, H., Stockwell, J., Jessulat, M., Goebels, F., Broderik, K., Scott, N.E., Vlasblom, J., Musso, G., Prasad, B., Lamantea, E., Garavaglia, B., Rajput, A., Murayama, K., Okazaki, Y., Foster, L.J., Bader, G.D., Cayabyab, F.S., Babu, M.*
iScience 2019;19:1114-1132. (*corresponding)
PMID:31536960
- A Gaussian process-based definition reveals new and bona fide genetic interactions compared to a multiplicative model in the Gram-negative Escherichia coli
Kumar, A., Hosseinnia, A., Gagarinova, A., Phanse, S., Kim, S., Aly, K.A., Zilles, S., Babu, M.*
Bioinformatics 2020;36(3):880-889. (*corresponding)
PMID:31504172
- A Tag-Based Affinity Purification Mass Spectrometry Workflow for Systematic Isolation of the Human Mitochondrial Protein Complexes.
Wu, Z., Malty, R., Moutaoufik, M.T., Zhang, Q., Jessulat, M., Babu, M.
Adv Exp Med Biol.2019;1158:83-100.
PMID:31452137
- Profiling the E. coli membrane interactome captured in peptidisc libraries
Carlson, M.L., Stacey, R.G., Young, J.W., Wason, I.S.,Zhao, Z., Rattray, D.G., Scott, N., Kerr, C.H., Babu, M., Foster, L.J., Duong F.V.H.
eLife 2019;8:e46615.
PMID:31364989
- Mitochondria in Health and in Sickness (1st ed. 2019 Edition)
Editors: Urbani, A & Babu, M.
Advances in Experimental Medicine and Biology 2019, Springer Publishing, New York
ISBN 978-9811383663
- Identification and Functional Testing of Novel Interacting Protein Partners for the Stress Sensors Wsc1p and Mid2p of Saccharomyces cerevisiae
Santiago-Cartagena, E., González-Crespo, S., Vélez, V., Martínez, N., Snider, J., M., Jessulat, Aoki, H., Minic, Z., Akamine, P., Mejías, I., Pérez, L.M., Rymond, B.C., Babu, M., Stagljar I., and Rodríguez-Medina, J.R.
G3 (Bethesda). 2019;9(4):1085-1102.
PMID:30733383
- In silico engineering of synthetic binding proteins from random amino acid sequences
Burnside, D., Schoenrock, A., Moteshareie, H., Hooshyar, M., Basra, P., Hajikarimlou, M., Dick, K., Barnes, B., Kazmirchuk, T., Jessulat, M., Pitre, S., Samanfar, B., Babu, M., Green, J.R., Wong, A., Dehne, F., Biggar, K.K., Golshani, A.
iScience 2019;11:375-387.
PMID:30660105
↑ Top
- Chromatographic separation strategies for precision mass spectrometry to study protein-protein interactions and protein phosphorylation
Minic, Z., Dahms, T.E.S., Babu, M.
Journal of Chromatography B, 2018;1102-1103:96-108.
PMID:30380468
- Heavy metal sensitivities of gene deletion strains for ITT1 and RPS1A connect their activities to the expression of URE2,a key gene involved in metal detoxification in yeast
Moteshareie, H., Hajikarimlou, M., Indrayanti, A.M., Burnside, D., Dias, A.P., Lettl, C., Ahmed, D., Omidi, K., Kazmirchuk, T., Puchacz, N., Zare, N., Takallou, S., Naing, T., Hernández, R.B., Willmore, W., Babu, M., McKay, B., Samanfar, B., Holcik, M., Golshani A.
PLoS One. 2018;13(9):e0198704.
PMID:30231023
- The uridylyltransferase, GlnD, and tRNA modification GTPase, MnmE allosterically control Escherichia coli folylpoly-γ-glutamate synthase, FolC.
Rodionova, I. A., Goodacre, N., Do, J., Hosseinnia, A., Babu, M., Uetz, P. and Saier Jr. M.H.
Journal of Biological Chemistry, 2018;293(40):15725-15732.
PMID:30089654
- Acyldepsipeptide Analogs Dysregulate Human Mitochondrial ClpP Protease Activity and Cause Apoptotic Cell Death
Wong, K. S., Mabanglo, M.F., Seraphim, T.V., Mollica, A., Mao, Y., Rizzolo, K., Leung, E., Moutaoufik, M. T., Hoell, L., Phanse, S., Goodreid, J., Barbosa, L.R.S., Ramos, C.H.I., Babu, M., Mennella, V., Batey, R.A., Schimmer, A.D., Houry, W.A.
Cell Chemical Biology. 2018;25(8):1017-1030.e9.
PMID:30126533
Preview
- EPHB6 augments both development and drug sensitivity of triple-negative breast cancer tumours
Toosi, B.M., El Zawily, A., Truitt, L., Shannon, M., Allonby, O., Babu, M., DeCoteau, J., Mousseau, D., Ali, M., Freywald, T., Gall, A.. Vizeacoumar, F. S., Kirzinger, M. W., Geyer, C.R., Anderson, D.H., Kim, T., Welm, A.L., Siegel, P., Vizeacoumar, F.J., Kusalik, A., & Freywald, A.
Oncogene 2018;37(30):4073-4093.
PMID:29700392
- Zinc oxide and silver nanoparticles toxicity in the baker's yeast, Saccharomyces cerevisiae.
Galván Márquez, I., Ghiyasvand, M., Massarsky, A., Babu, M., Samanfar, B., Omidi, K., Moon, T. W., Smith, M. L., Golshani, A.
PLoS One. 2018;13(3):e0193111.
PMID:29554091
- Systems analysis of the genetic interaction network of yeast molecular chaperones
Rizzolo, K., Kumar, A., Kakihara, Y., Phanse, S., Minic, Z., Snider, J., Stagljar, I., Zilles, S., Babu, M.* and Houry, W. A.*
Molecular Omics. 2018;14(2):82-94. (*co-corresponding)
PMID:29659649
- Metabolic adaptation of a C-terminal protease A-deficient Rhizobium leguminosarum in response to loss of nutrient transport
Jun, D., Minic, Z., Bhat, S.V., Vanderlinde, L.M., Yost, C.K., Babu, M. and Dahms, T.E.
Frontiers in Microbiology 2018;8:2617.
PMID:29354107
- Computational Analysis of the Chaperone Interaction Networks.
Kumar. A., Rizzolo. K., Zilles. S., Babu. M., Houry, W.
Methods in Molecular Biology 2018;1709:275-291. (*co-corresponding)
PMID:29177666
- Global landscape of cell envelope protein complexes in Escherichia coli
Babu, M.*, Bundalovic-Torma, C., Calmettes, C., Phanse, S., Zhang, Q., Jiang, Y., Minic, Z., Kim, S., Mehla, J., Gagarinova, A., Rodionova, I., Kumar, A., Guo, H., Kagan, O., Pogoutse, O., Aoki, H., Deineko, V., Caufield, J. H., Holtzapple, E., Zhang, Z., Vastermark, A., Pandya, Y., Lai, C. C., El Bakkouri, M., Hooda, Y., Shah, M., Burnside, D., Hooshyar, M., Vlasblom, J., Rajagopala, S.V., Golshani, A., Wuchty, S., Greenblatt, J.F., Saier, M., Uetz, P., F Moraes, T., Parkinson, J., Emili, A.
Nature Biotechnology 2018;36(1):103-112. (*corresponding)
PMID:29176613
Preview
- Uncharacterized ORF HUR1 influences the efficiency of non-homologous end-joining repair in Saccharomyces cerevisiae
Omidi, K., Jessulat, M., Hooshyar, M., Burnside, D., Schoenrock, A., Kazmirchuk, T., Hajikarimlou, M., Daniel, M., Moteshareie, H., Bhojoo, U., Sanders, M., Ramotar, D., Dehne, F., Samanfar, B., Babu, M., Golshani, A.
Gene, 2018;639:128-136.
PMID:28987344
- The nitrogen regulatory PII protein (GlnB) and N-acetyl-glucosamine 6-phosphate epimerase (NanE) allosterically activate glucosamine 6-phosphate deaminase (NagB) in Escherichia coli.
Rodionova, I.A., Goodacre, N., Babu, M., Emili, A., Uetz, P., and Saier Jr., M. H.
Journal of Bacteriology 2018;200(5):e00691-17.
PMID:29229699
↑ Top
- The sensitivity of the yeast, Saccharomyces cerevisiae, to acetic acid is influenced by DOM34 and RPL36A
Samanfar, B., Shostak, K., Moteshareie, H., Hajikarimlou, M., Shaikho, S., Omidi, K., Hooshyar, M., Burnside, D., Márquez, I.G., Kazmirchuk, T., Naing, T., Ludovico, P., York-Lyon, A., Szereszewski, K., Leung, C., Jin, J.Y., Megarbane, R., Smith, M.L., Babu, M., Holcik, M., Golshani, A.
PeerJ 2017;5:e4037.
PMID:29158977
- Renal oncocytoma characterized by the defective complex I of the respiratory chain boosts the synthesis of the ROS scavenger glutathione
Kürschner, G., Zhang, Z., Clima, R., Xiao, Y., Busch, J.F., Kilic, E., Jung, K., Berndt, N., Bulik, S., Holzhütter, H-G., Gasparre, G., Attimonelli, M., Babu, M.*, and Meierhofer, M.
Oncotarget 2017;8(62):105882-105904. (*co-corresponding)
PMID:29285300
- A map of human mitochondrial protein interactions linked to neurodegeneration reveals new mechanisms of redox homeostasis and NF-κB signaling
Malty, R. H., Aoki, H., A., K., Phanse, S., Amin, S., Zhang, Q., Minic, Z., Goebels, F., Musso, G., Wu, Z., Abou-tok, H., Meyer, M., V., D., Kassir, S., Sidhu, V., Jessulat, M., Scott, N. E., Xiong, X., Vlasblom, J., Prasad, B., Foster, L. J., Alberio, T., Garavaglia, B., Yu, H., Bader, G. D., Nakamura, K., Parkinson, J., and Babu, M*.
Cell Systems 2017;5(6):564-577.e12. (*corresponding)
PMID:29128334
Preview
- Designing anti-Zika virus peptides derived from predicted human-Zika virus protein-protein interactions
Kazmirchuk, T., Dick, K., Burnside, D.J., Barnes, B., Moteshareie, H., Hajikarimlou, M., Omidi, K., Ahmed, D., Low, A., Lettl, C., Hooshyar, M., Schoenrock, A., Pitre, S., Babu, M., Cassol, E., Samanfar, B., Wong, A., Dehne, F., Green, J.R. Golshani, A.
Computational Biology and Chemistry 2017;71:180-187
PMID:29112936
- Features of the Chaperone Cellular Network Revealed through Systematic Interaction Mapping
Rizzolo, K., Huen, J., Kumar, A., Phanse, S., Vlasblom, J., Kakihara,Y., Zeineddine, H. A., Minic, Z., Snider, J., Wang, W., Pons, C., Seraphim, T. V., Boczek, E. E., Alberti, S., Costanzo, M., Myers, C. L., Stagljar, I., Boone, C., Babu*, M., Houry*, W. A.
Cell Reports 2017;20(11):2735-2748. (*co-corresponding)
PMID:28903051
- Insights from protein-protein interaction studies on bacterial pathogenesis
Gagarinova, A., Phanse, S., Cygler, M. & Babu*, M.
Expert Review of Proteomics 2017;14(9):779-797. (*corresponding)
PMID:28786313
- HIV-1 Gp120 clade B/C induces a GRP78 driven cytoprotective mechanism in astrocytoma
López, S.N., Rodríguez-Valentín, M., Rivera, M., Rodríguez, M., Babu, M., Cubano, L.A., Xiong, H., Wang, G., Kucheryavykh, L., and Boukli, N.M.
Oncotarget 2017;8(40):68415-68438.
PMID:28978127
- The phosphocarrier protein HPr of the bacterial phosphotransferase system globally regulates energy metabolism by directly interacting with multiple enzymes in Escherichia coli.
Rodionova, I.A., Zhang, Z., Mehla, J., Goodacre, N., Babu, M., Emili, A., Uetz, P., Saier, M.H. Jr.
Journal of Biological Chemistry 2017;292(34):14250-14257.
PMID:28634232
- Systematic protein–protein interaction mapping for clinically relevant human GPCRs
Sokolina, K., Kittanakom, S., Snider, J., Kotlyar, M., Maurice, P., Gandía, J., Benleulmi-Chaachoua, A., Tadagaki, K., Oishi, A., Wong, V., Malty, R.H., Deineko, V., Aoki, H., Amin, S., Yao, Z., Morató, X., Otasek, D., Kobayashi, H., Menendez, J., Auerbach, D., Angers, S., Pržulj, N., Bouvier, M., Babu, M., Ciruela, F., Jockers, R., Jurisica, I., Stagljar, I.
Molecular Systems Biology 2017;13(3):918.
PMID:28298427
- A Global Analysis of the Receptor Tyrosine Kinase-Protein Phosphatase Interactome
Yao, Z., Darowski, K., St-Denis, N., Wong, V., Offensperger, F., Villedieu, A., Amin, S., Malty, R., Aoki, H., Guo, H., Xu, Y., Iorio, C., Kotlyar, M., Emili, A., Jurisica, I., Neel, B.G., Babu, M., Gingras, A., Stagljar I.
Molecular Cell 2017;65(2):347-360.
PMID:28065597
↑ Top
- Systematic Genetic Screens Reveal the Dynamic Global Functional Organization of the Bacterial Translation Machinery
Gagarinova, A., Stewart, G., Samanfar, B., Phanse, S., White, C.A., Aoki, H., Deineko, V., Beloglazova, N., Yakunin, A.F., Golshani, A., Brown, E.D., Babu, M., Emili, A.
Cell Reports 2016;17(3):904-916.
PMID:27732863
- Novel Interactome of Saccharomyces cerevisiae Myosin Type II Identified by a Modified Integrated Membrane Yeast Two-Hybrid (iMYTH) Screen.
Santiago, E., Akamine, P., Snider, J., Wong, V., Jessulat, M., Deineko, V., Gagarinova, A., Aoki, H., Minic, Z., Phanse, S., San Antonio, A., Cubano, L., Rymond, B.C., Babu, M., Stagljar, I., Rodriguez-Medina, J.R.
G3 (Bethesda) 2016;6(5):1469-74,
PMID:26921299
- Towards a functional definition of the mitochondrial human proteome
Fasano, M., Alberio, T., Babu, M., Lundberg, E., Urbani, A.
EuPA Open Proteomics 2016;10:24-27.
PMID:29900096
- Conditional Epistatic Interaction Maps Reveal Global Functional Rewiring of Genome Integrity Pathways in Escherichia coli.
Kumar, A., Beloglazova, N., Bundalovic-Torma, C., Phanse, S., Deineko, V., Gagarinova, A., Musso, G., Vlasblom, J., Lemak, S., Hooshyar, M., Minic, Z., Wagih, O., Mosca, R., Aloy, P., Golshani, A., Parkinson, J., Emili, A., Yakunin A.E., and Babu M.*
Cell Reports 2016;14(3):648-661. (*corresponding)
PMID:26774489
↑ Top
- Proteome-wide dataset supporting the study of ancient metazoan macromolecular complexes
Phanse, S., Wan, C., Borgeson, B., Tu, F., Drew, K., Clark, G., Xiong, X., Kagan, O., Kwan, J., Bezginov, A., Chessman, K., Pal, S., Cromar, G., Papoulas, O., Ni, Z., Boutz, D.R., Stoilova, S., Havugimana, P.C., Guo, X., Malty, R.H., Sarov, M., Greenblatt, J., Babu, M., Derry, W.B., Tillier, E.R., Wallingford, J.B., Parkinson, J., Marcotte, E.M. & Emili, A.
Data in Brief, 2015;6:715-21.
PMID:26870755
- Identification of Human Neuronal Protein Complexes Reveals Biochemical Activities and Convergent Mechanisms of Action in Autism Spectrum Disorders
Li, J., Ma, Z., Shi, M., Malty, R.H., Aoki, H., Minic, Z., Phanse, S., Jin, K., Wall, D.P., Zhang, Z., Urban, A.E., Hallmayer, J., Babu, M*., Snyder, M*.
Cell Systems 2015;1(5):361-374. (*co-corresponding)
PMID:26949739
Preview
- Panorama of ancient metazoan macromolecular complexes
Wan, C., Borgeson, B., Phanse, S., Tu, F., Drew, K., Clark, G., Xiong, X., Kagan, O., Kwan, J.,Bezginov, A., Chessman, K., Pal, S., Cromar, G., Papoulas, O., Ni, Z., Boutz, D. R., Stoilova, S., Havugimana, P. C., Guo, X., Malty, R. H., Sarov, M., Greenblatt, J., Babu, M., Derry, W. B., Tillier, E. R., Wallingford, J. B., Parkinson, J., Marcotte, E. M. & Emili, A.
Nature 2015;525(7569):339-44.
PMID: 26344197
Preview
- Quantitative and Systems-Based Approaches for Deciphering Bacterial Membrane Interactome and Gene Function
Deineko, V., Kumar, A., Vlasblom, J., Babu, M*.
Advances in Experimental Medicine and Biology 2015;883:135-54. (*corresponding)
PMID: 26621466
- Prokaryotic Systems Biology(1st ed).
Editors: Krogan, N. J.,& Babu, M.
Advances in Experimental Medicine and Biology 2015, Springer Publishing, New York
ISBN 978-3-319-23602-5
- Functional Diversity of Haloacid Dehalogenase Superfamily Phosphatases from Saccharomyces cerevisiae: Biochemical, Structural, and Evolutionary Insights
Kuznetsova, E., Nocek2, B., Brown, G., Makarova, K, S,. Flick, R., Wolf, Y. I., Khusnutdinova, A., Evdokimova, E., Jin, K., Tan, K., Hanson, A. D., Hasnain, G., Zallot, R., de Crecy-Lagard, V., Babu, M., Savchenko, A., Joachimiak, A., Edwards, A. M., Koonin, E. V. and Yakunin, A. M.
The Journal of Biological Chemistry 2015;290(30):18678-98.
PMID: 26071590
- Spindle Checkpoint Factors Bub1 and Bub2 Promote DNA Double Strand Break Repair by Non-Homologous End Joining
Jessulat, M., Malty, R., Nguyen-Tran, D., Deineko, V., Aoki, H., Vlasblom, J., Omidi, K., Jin, K., Minic, Z., Hooshyar, M., Burnside, D., Samanfar, B., Phanse, S., Freywald, T., Prasad, B., Zhang, Z., Vizeacoumar, F., Krogan, N., Freywald, A., Golshani, A., and Babu, M*.
Molecular and Cellular Biology 2015;35(14):2448-63. (*corresponding)
PMID: 25963654
- A comprehensive membrane interactome mapping of Sho1p reveals Fps1p as a novel key player in the regulation of the HOG pathway in S. cerevisiae.
Lam, M.H.Y., Snider, J., Rehal, M., Wong, V., Aboualizadeh, F., Drecun, L., Wong, O., Jubran, B., Li, M., Ali, M., Jessulat, M., Dieneko, V., Miller, R., Lee, M.E., Park, H.O., Davidson, A., Babu, M. and Stagljar, I.
Journal of Molecular Biology 2015;427(11):2088-103.
PMID: 25644660
- Rab5 family guanine nucleotide exchange factors bind retromer and promote its recruitment to endosomes.
Bean, D.M.B., Davey, M., Snider, J., Jessulat, M., Dieneko, V., Tinney, M., Staglijar, I., Babu, M., Conibear, E.
Molecular Biology of the Cell 2015;26(6):1119-28
PMID: 25609093
- Yeast mitochondrial protein-protein interactions reveal diverse complexes and disease-relevant functional relationships
Jin, K., Musso, G., Vlasblom, J., Jessulat, M., Deineko, V., Negroni, J., Mosca, R., Malty, R., Nguyen-Tran, D-H., Aoki, H., Minic, Z., Freywald, T., Phanse, S., Xiang, Q., Freywald, A., Aloy, P., Zhang, Z and Babu, M*
Journal of Proteome Research 2015;14(2):1220-37 (*corresponding)
PMID: 25546499
- Novel function discovery with GeneMANIA: a new integrated resource for gene function prediction in Escherichia coli
Vlasblom, J., Zuberi, K., Rodriguez, H., Arnold, R., , Gagarinova, A., Deineko, V., Kumar, A., Leung, E., Rizzolo, K., Samanfar, B., Chang, L., Phanse, S., Golshani, A., Greenblatt, J.F., Houry, W.A., Emili, A., Morris, Q., Bader, G., and Babu, M.*
Bioinformatics 2015;31(3):306-10. (*corresponding)
PMID: 25316676
- Mitochondrial Targets for Pharmacological Intervention in Human Disease
Malty, R.H., Jesssulat, M., Jin, K., Musso, G., Vlasblom, J., Phanse, S., Zhang, Z., Babu, M.*
Journal of Proteome Research 2015;14(1):5-21. (*corresponding)
PMID: 25367773
↑ Top
- Efficient Prediction of Human Protein-protein Interactions at a Global Scale
Schoenrock, A., Samanfar, B., Pitre, S., Hooshyar, M., Jin, K., Phillips, C., Wang, H., Phanse, S., Omidi, K., Gui, Y., Alamgir, Md., Wong, A., Barrenas, Fredrik., Babu, M., Benson, M., Langston, M., Green, J.R., Dehne, F., Golshani, A.
BMC Bioinformatics 2014;15(1):383. [Epub ahead of print]
PMID: 25492630
- Ligand stimulation induces clathrin- and Rab5- dependent down regulation of the kinase-dead EphB6 receptor preceded by the disruption of EphB6-Hsp90 interaction
Allonby, O., El Zawily, A.M., Freywald, T., Mousseau, D.D., Chlan, J., Anderson, D., Benmerah, A., Sidhu, V., Babu, M., DeCoteau, J. and Freywald, A.
Cellular Signalling 2014;12:2645 - 2657.
PMID: 25152371
- Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli
Babu, M*, Arnold, A., Bundalovic-Torma, C., Gagarinova, A., Wong, K.S., Kumar, A., Stewart, G., Samanfar, B., Aoki, H., Wagih, O., Vlasblom, J., Phanse, S., Lad, K., Yu, AYH, Graham, C., Jin, K., Brown, E., Golshani, A., Kim, P., Moreno-Hagelsieb, G., Greenblatt, J., Houry, W.A., Parkinson, J. and Emili, A.
PLoS Genetics 2014;10(2):e1004120 (*corresponding)
PMID: 24586182
- Binary Protein-protein Interaction Landscape of Escherichia coli
Rajagopala, S.V*., Sikorski, P., Kumar, A., Mosca, R., Vlasblom, J., Arnold, R., Franca-Koh, J., Pakala, S.B., Phanse, S., Ceol, A., Häuser, R., Siszler, G., Wuchty, S., Emili, A., Babu, M*., Aloy, P., Pieper, R. and Uetz, P.
Nature Biotechnology 2014;32:285-290. (*co-corresponding)
PMID: 24561554
Preview
- A global investigation of gene deletion strains that affect premature stop codon bypass in yeast, Saccharomyces cerevisiae
Samanfar, B., Tan, L.H., Shostak, K., Chalabian, F., Wu, Z., Alamgir MD, Sunba, B., Burnside, D., Omidi, K., Hooshyar, M., Márquez, I.G., Jessulat, M., Smith, M.L., Babu, M., Azizid, A. and Golshani, A.
Molecular BioSystems 2014;10:916-924
PMID: 24535059
- Phosphatase Complex Pph3/Psy2 Is Involved in Regulation of Efficient Non-Homologous End-Joining Pathway in the Yeast Saccharomyces cerevisiae.
Omidi, K., Hooshyar, M., Jessulat, M., Samanfar, B., Sanders, M., Burnside, D., Pitre, S., Schoenrock, A., Xu, J., Babu, M. and Golshani, A.
PLoS One 2014;9(1):e87248
PMID: 24498054
- The MoxR ATPase RavA and its cofactor ViaA interact with the NADH:ubiquinone oxidoreductase I in Escherichia coli
Wong, K.S., Snider, J.D., Graham, C., Greenblatt, J.F., Emili, A., Babu, M. and Houry, W.A.
PLoS One 2014;9(1):e85529.
PMID: 24454883
- NleH defines a new family of bacterial effector kinases
Grishin, A.M., Cherney, M., Anderson, D.H., Phanse, S., Babu, M. and Cygler, M.
Structure 2014;2:250 - 259.
PMID: 24373767
- Exploring Mitochondrial Systems Properties of Neurodegenerative Diseases through Interactome Mapping
Vlasblom J., Jin K., Kassir S., Babu M.*
Journal of Proteomics 2014;100:8 - 24. (* corresponding)
PMID: 24262152
↑ Top
- A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities
Vizeacoumar, F. J., Arnold, R., Vizeacoumar, F. S., Chandrashekhar, M., Buzina, A., Young, J. T. F., Kwan, J.H.M., Sayad, A., Mero, P., Lawo, S., Tanaka, H., Brown, K. R., Baryshnikova, A., Mak, A. B., Fedyshyn, Y., Wang, Y., Brito, G. C., Kasimer, D., Makhnevych, T., Ketela, T., Datti, A., Babu, M., Emili, A., Pelletier L, Wrana J, Wainberg Z, Kim P. M., Rottapel R, O'Brien C. A., Andrews B, Boone C, Moffat J.
Molecular Systems Biology 2013;9:696
PMID: 24104479
- Investigating the Ustilago maydis/Zea mays pathosystem: transcriptional responses and novel functional aspects of a fungal calcineurin regulatory B subunit
Donaldson, M. E., Meng, S., Gagarinova, A., Babu, M., Lambie, S. C., Swiadek, A. A., Saville, B. J.
Fungal Genetics and Biology 2013;59:91-104
PMID: 23973481
- ER exit sites are physical and functional core autophagosome biogenesis components
Graef, M., Freidman, J., Graham, C., Babu, M., Nunnari, J.
Molecular Biology of the Cell 2013;24:2918-2931.
PMID: 23904270
- Mapping the functional yeast ABC transporter interactome.
Snider, J., Hanif, A. Lee, M. E., Jin, K., Yu, Q. R., Graham, C., Chuk, M., Damjanovic, D., Wierzbicka, M., Tang, P., Balderes, D., Wong, V., Jessulat, M., Darowski, K. D., San Luis, B., Shevelev, I., Sturley, S. L., Boone, C., Greenblatt, J. F., Zhang, Z., Paumi, C. M., Babu, M., Park, H., Michaelis S., & Stagljar, I.
Nature Chemical Biology 2013;9:565-572.
PMID: 23831759
Preview
↑ Top
- Identification of protein complexes in Escherichia coli using sequential peptide affinity purification in combination with tandem mass spectrometry.
Babu, M., Kagan, O., Guo, H., Greenblatt, J. and Emili, A.
Journal of Visualized Experiments 2012;(69): 4057
PMID: 23168686
- Mapping bacterial functional networks and pathways in Escherichia coli using synthetic genetic arrays.
Gagarinova, A., Babu, M., Greenblatt, J. and Emili, A.
Journal of Visualized Experiments 2012;(69): 4056.
PMID: 23168417
-
- Interaction landscape of membrane protein complexes in Saccharomyces Cerevisiae.
Babu, M., Vlasblom, J., Pu, S., Guo, X., Graham, C., Vizeacoumar, F.J., Bean, B.D.M., Phanse, S., Fong, V., Davey, M., Snider, J., Hnatshak, O., Bajaj, N., Chandran, S., Punna, T., Christopolous, C., Wong, V., Zhong, G., Li, J., Stagljar, I., Conibear, E., Wodak, S.J.W., Emili, A. and Greenblatt, J.F.
Nature 2012;489:585-9
PMID: 22940862
Preview
-
- A Census of Human Soluble Protein Complexes.
Havugimana, P.C., Hart, G.T., Nepusz, T., Yang, H., Turinsky, A.L., Li, Z., Wang, P.I., Boutz, D.R., Fong, V., Phanse, S., Babu, M., Craig, S.A., Hu, P., Wan, C., Vlasblom, J., Dar, V., Bezginov, A., Clark, G.W., Wu, G.C., Wodak, S.J., Tillier, E.R.M., Paccanaro, A., Marcotte, E.M., Emili, A.
Cell 2012;150:1068-1081
PMID: 22939629
Preview
-
↑ Top
- Genetic interaction maps in Escherichia coli reveal condition-dependent functional crosstalk among cell envelope biogenesis pathways.
Babu, M*., Díaz-Mejía, J.J., Vlasblom, J., Gagarinova, A., Phanse, S., Graham, C., Arnold, R., Yousif, F., Ding, H., Xiong, X., Nazarians-Armavil, A., Alamgir, Md., Ali, M., Pogoutse, O., Pe′er, A., Parkinson, J., Golshani, A., Whitfield, C., Wodak, S.J., Moreno-Hagelsieb, G., Greenblatt, J.F. and Emili, A.*
PLoS Genetics 2011;7:e1002377. (*co-corresponding)
PMID: 22125496
- Array-based synthetic genetic screens to map bacterial pathways and functional networks in Escherichia coli.
Babu, M., Gagarinova, A., Greenblatt, J.F. and Emili, A
Methods Molecular Biology 2011;781:99-126.
PMID: 21877280
- Ribosome-dependent ATPase interacts with conserved membrane protein in Escherichia coli to modulate protein synthesis and oxidative phosphorylation.
Babu, M., Aoki, H., Chowdhury, W.Q., Gagarinova, A., Graham, C., Laliberte, B., Hasan, N., Xu, J., Golshani, A., Emili, A., Greenblatt, J. and Ganoza, M.C.
PLoS One 2011;6:e18510.
PMID: 21556145
- A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Babu, M.*, Beloglazova, N.*, Flick, R., Graham, C., Skarina, T., Nocek, B., Gagarinova, A., Pogoutse, O., Brown, G., Binkowski, A., Phanse, S., Joachimiak, A., Koonin, E. V., Savchenko, A., Emili, A., Greenblatt, J., Edwards, A. M., Yakunin, A. F.
Molecular Microbiology 2011;79:484-502. (*co-first author)
PMID: 21219465
- Array-based synthetic genetic screens to map bacterial pathways and functional networks in Escherichia coli.
Babu, M., Gagarinova, A., Greenblatt, J.F. and Emili, A
Methods Molecular Biology 2011;765:125-153.
PMID: 21815091
↑ Top
- Quantifying the Escherichia coli proteome and transcriptome in a single cell with single-molecule sensitivity
Taniguchi, Y., Choi, P.J., Li, G-W., Chen, H., Babu, M., Hearn, J., Emili, A. and Xie, X.S.
Science 2010;329:533-538.
PMID: 20671182
Preview
- Structure of a SLC26 anion transporter STAS domain in complex with acyl carrier protein: implications for E. coli YchM in fatty acid metabolism.
Babu, M., Greenblatt, J., Emili, A., Strynadka, N. C.J., Reithmeier, A.F. and Moraes, T.F.
Structure 2010;18:1450-1462.
PMID: 21070944
↑ Top
- Systems-level approaches for identifying and analyzing genetic interaction networks in Escherichia coli and extensions to other prokaryotes.
Babu, M.*, Musso, G.*, Diaz-Mejia, J.J.*, Butland, G., Greenblatt, J. and Emili, A.
Molecular BioSystems 2009;5:1439-1455. (*co-first author)
PMID: 19763343
- Sequential peptide affinity purification system for the systematic isolation and identification of protein complexes from Escherichia coli.
Babu, M., Butland, G., Pogoutse, O., Li, J., Greenblatt, J.F. and Emili, A.
Methods Molecular Biology 2009;564:373-400.
PMID: 19544035
- Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins.
Hu, P.*, Janga, S.C.*, Babu, M.*, Diaz-Mejia, J.J.*, Butland, G.*, Yang, W., Pogoutse, O., Guo, X., Phanse, S., Wong, P., Chandran, S., Christopoulos, C., Nazarians-Armavi,l A., Nasseri, N. K., Musso, G., Ali, M., Nazemof, N., Eroukova, V., Golshani, A., Paccanaro, A., Greenblatt, J. F., Moreno-Hagelsieb, G., Emili, A.
PLoS Biology 2009;7:e96. (*co-first author)
PMID: 19402753
Preview
- Computational and experimental approaches to chart the Escherichia coli cell-envelope associated proteome and interactome.
Diaz-Mejia, J.J.*, Babu, M.* and Emili, A.
FEMS Microbiology Reviews 2009;33:66-97. (*co-first author)
PMID: 19054114
- Systematic characterization of the protein interaction network and protein complexes in Saccharomyces cerevisiae using tandem affinity purification and mass spectrometry
Babu, M., Krogan, N.J., Awrey, D.E., Emili, A. and Greenblatt, J.F.
Methods Molecular Biology 2009;548:187-207.
PMID: 19521826
↑ Top
- eSGA: E. coli synthetic genetic array analysis.
Butland, G.*, Babu, M.*, Diaz-Mejia, J.J., Bohdana, F., Phanse, S., Gold, B., Yang, W., Li, J., Gagarinova, A. G., Pogoutse, O., Mori, H., Wanner, B. L., Lo, H., Wasniewski, J., Christopolous, C., Ali, M., Venn, P., Safavi-Naini, A., Sourour, N., Caron, S., Choi, J. Y., Laigle, L., Nazarians-Armavil, A., Deshpande, A., Joe, S., Datsenko, K. A., Yamamoto, N., Andrews, B. J., Boone, C., Ding, H., Sheikh, B., Moreno-Hagelseib, G., Greenblatt, J. F., Emili, A.
Nature Methods 2008;5:789-795. (*co-first author)
PMID: 18677321
Preview
- Recombination analysis of soybean mosaic virus sequences reveals evidence of RNA recombination between distinct pathotypes.
Gagarinova, A.G., Babu, M., Strömvik, M.V. and Wang, A.
Virology J. 2008;5:143.
PMID: 19036160
- Identification and molecular characterization of two naturally occurring soybean mosaic virus isolates that are closely related but differ in their ability to overcome Rsv4 resistance.
Gagarinova, A.G., Babu, M., Poysa, V., Hill, J.H. and Wang, A.
Virus Research 2008;138:50-56.
PMID: 18793685
- The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves.
Tang, X., Hou, A., Babu, M., Nguyen, V., Hurtado, L., Lu, Q., Reyes, J., Wang, A., Keller, W.A., Tsang, E.D. and Cui, Y.
Plant Physiology 2008;147:1143-1157.
PMID: 18508955
- Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection.
Babu, M., Griffiths, J.S., Huang, T.S., and Wang A.
BMC Genomics 2008;9:325.
PMID: 18613973
- Association of the transcriptional response of soybean plants with soybean mosaic virus systemic infection.
Babu, M., Gagarinova, A.G., Brandle, J.E. and Wang, A.
Journal General Virology 2008;89:1069-1080.
PMID: 18343851
↑ Top
- Differential gene expression in filamentous cells of Ustilago maydis.
Babu, M., Choffe, K. and Saville, B.J.
Current Genetics 2005;47:316-333.
PMID: 15809875
- Differential gene expression during Ustilago maydis teliospore germination..
Zahiri, A.R., Babu, M. and Saville, B.J.
Molecular Genetics Genomics 2005;273:394-403.
PMID: 15887033
↑ Top
- A comparative genomic analysis of ESTs from Ustilago maydis.
Austin, R., Provart, N. J., Sacadura, N.T., Nugent, K.G., Babu, M. and Saville, B. J.
Functional and Integrative Genomics 2004;4:207-218.
PMID: 15349794
↑ Top










































